| Full name: membrane spanning 4-domains A14 | Alias Symbol: NYD-SP21|FLJ32856|DKFZp434H092 | ||
| Type: protein-coding gene | Cytoband: 11q12.2 | ||
| Entrez ID: 84689 | HGNC ID: HGNC:30706 | Ensembl Gene: | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of MS4A14:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MS4A14 | 84689 | 229510_at | 0.2811 | 0.4428 | |
| GSE26886 | MS4A14 | 84689 | 229510_at | -0.0129 | 0.9190 | |
| GSE45670 | MS4A14 | 84689 | 229510_at | 0.0943 | 0.6794 | |
| GSE63941 | MS4A14 | 84689 | 229510_at | 0.1319 | 0.2438 | |
| GSE77861 | MS4A14 | 84689 | 229510_at | 0.0489 | 0.5116 | |
| GSE97050 | MS4A14 | 84689 | A_33_P3290577 | 0.4911 | 0.2318 | |
| SRP064894 | MS4A14 | 84689 | RNAseq | 1.0673 | 0.0036 | |
| SRP133303 | MS4A14 | 84689 | RNAseq | 0.6539 | 0.0663 | |
| SRP159526 | MS4A14 | 84689 | RNAseq | -0.1071 | 0.7782 | |
| SRP219564 | MS4A14 | 84689 | RNAseq | 1.0984 | 0.0523 | |
| TCGA | MS4A14 | 84689 | RNAseq | 0.1896 | 0.4097 |
Upregulated datasets: 1; Downregulated datasets: 0.
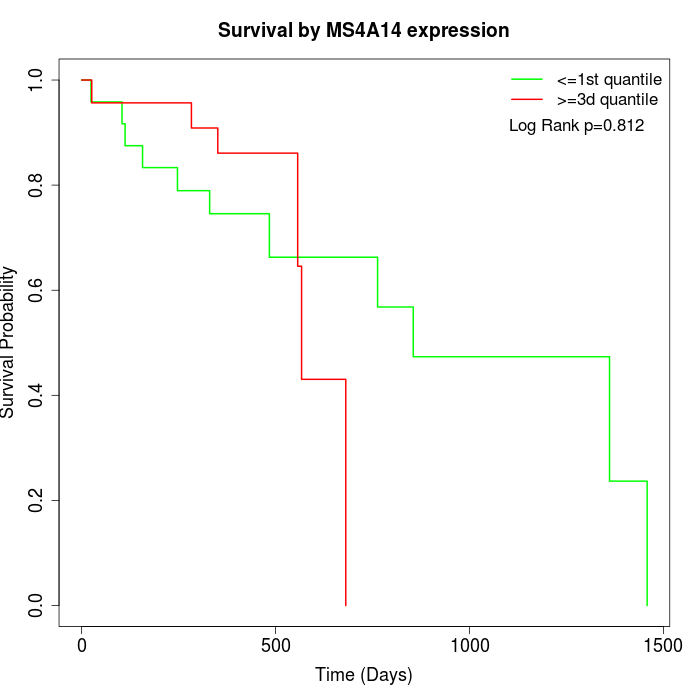
Survival by MS4A14 expression:
Note: Click image to view full size file.
Copy number change of MS4A14:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MS4A14 | 84689 | 2 | 4 | 24 | |
| GSE20123 | MS4A14 | 84689 | 2 | 4 | 24 | |
| GSE43470 | MS4A14 | 84689 | 2 | 5 | 36 | |
| GSE46452 | MS4A14 | 84689 | 8 | 4 | 47 | |
| GSE47630 | MS4A14 | 84689 | 3 | 8 | 29 | |
| GSE54993 | MS4A14 | 84689 | 3 | 0 | 67 | |
| GSE54994 | MS4A14 | 84689 | 5 | 5 | 43 | |
| GSE60625 | MS4A14 | 84689 | 0 | 3 | 8 | |
| GSE74703 | MS4A14 | 84689 | 2 | 3 | 31 | |
| GSE74704 | MS4A14 | 84689 | 2 | 3 | 15 | |
| TCGA | MS4A14 | 84689 | 14 | 10 | 72 |
Total number of gains: 43; Total number of losses: 49; Total Number of normals: 396.
Somatic mutations of MS4A14:
Generating mutation plots.
Highly correlated genes for MS4A14:
Showing top 20/96 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MS4A14 | ITGAX | 0.757527 | 3 | 0 | 3 |
| MS4A14 | PSAP | 0.754333 | 3 | 0 | 3 |
| MS4A14 | PSTPIP2 | 0.73335 | 3 | 0 | 3 |
| MS4A14 | CD248 | 0.71583 | 3 | 0 | 3 |
| MS4A14 | TRPV2 | 0.712705 | 3 | 0 | 3 |
| MS4A14 | HERC5 | 0.706154 | 3 | 0 | 3 |
| MS4A14 | LCP2 | 0.699897 | 3 | 0 | 3 |
| MS4A14 | CMTM3 | 0.681473 | 3 | 0 | 3 |
| MS4A14 | C1QB | 0.680275 | 4 | 0 | 3 |
| MS4A14 | PUS3 | 0.672976 | 3 | 0 | 3 |
| MS4A14 | CTSZ | 0.671049 | 3 | 0 | 3 |
| MS4A14 | PREX1 | 0.670898 | 3 | 0 | 3 |
| MS4A14 | MS4A7 | 0.669449 | 4 | 0 | 4 |
| MS4A14 | GLIPR1 | 0.668818 | 3 | 0 | 3 |
| MS4A14 | WDR12 | 0.663843 | 3 | 0 | 3 |
| MS4A14 | TAP1 | 0.65767 | 4 | 0 | 3 |
| MS4A14 | C5orf15 | 0.656979 | 3 | 0 | 3 |
| MS4A14 | C1QA | 0.650461 | 4 | 0 | 4 |
| MS4A14 | TNFRSF1B | 0.649408 | 4 | 0 | 3 |
| MS4A14 | LILRB1 | 0.64683 | 5 | 0 | 4 |
For details and further investigation, click here