| Full name: integrin subunit alpha X | Alias Symbol: CD11c | ||
| Type: protein-coding gene | Cytoband: 16p11.2 | ||
| Entrez ID: 3687 | HGNC ID: HGNC:6152 | Ensembl Gene: ENSG00000140678 | OMIM ID: 151510 |
| Drug and gene relationship at DGIdb | |||
ITGAX involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04810 | Regulation of actin cytoskeleton | |
| hsa05152 | Tuberculosis |
Expression of ITGAX:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ITGAX | 3687 | 210184_at | 0.9526 | 0.1548 | |
| GSE20347 | ITGAX | 3687 | 210184_at | 0.2692 | 0.0138 | |
| GSE23400 | ITGAX | 3687 | 210184_at | 0.1538 | 0.0020 | |
| GSE26886 | ITGAX | 3687 | 210184_at | 0.2197 | 0.2493 | |
| GSE29001 | ITGAX | 3687 | 210184_at | 0.2836 | 0.1495 | |
| GSE38129 | ITGAX | 3687 | 210184_at | 0.2879 | 0.0107 | |
| GSE45670 | ITGAX | 3687 | 210184_at | 0.3855 | 0.0145 | |
| GSE53622 | ITGAX | 3687 | 61613 | 1.2101 | 0.0000 | |
| GSE53624 | ITGAX | 3687 | 61613 | 1.4721 | 0.0000 | |
| GSE63941 | ITGAX | 3687 | 210184_at | 0.1940 | 0.4528 | |
| GSE77861 | ITGAX | 3687 | 210184_at | 0.1436 | 0.3565 | |
| GSE97050 | ITGAX | 3687 | A_33_P3213169 | 0.7185 | 0.1736 | |
| SRP007169 | ITGAX | 3687 | RNAseq | 3.2552 | 0.0001 | |
| SRP008496 | ITGAX | 3687 | RNAseq | 1.9702 | 0.0005 | |
| SRP064894 | ITGAX | 3687 | RNAseq | 1.6323 | 0.0000 | |
| SRP133303 | ITGAX | 3687 | RNAseq | 1.2327 | 0.0000 | |
| SRP159526 | ITGAX | 3687 | RNAseq | 0.3565 | 0.5023 | |
| SRP193095 | ITGAX | 3687 | RNAseq | 1.6401 | 0.0000 | |
| SRP219564 | ITGAX | 3687 | RNAseq | 1.2021 | 0.0578 | |
| TCGA | ITGAX | 3687 | RNAseq | 0.6894 | 0.0000 |
Upregulated datasets: 7; Downregulated datasets: 0.
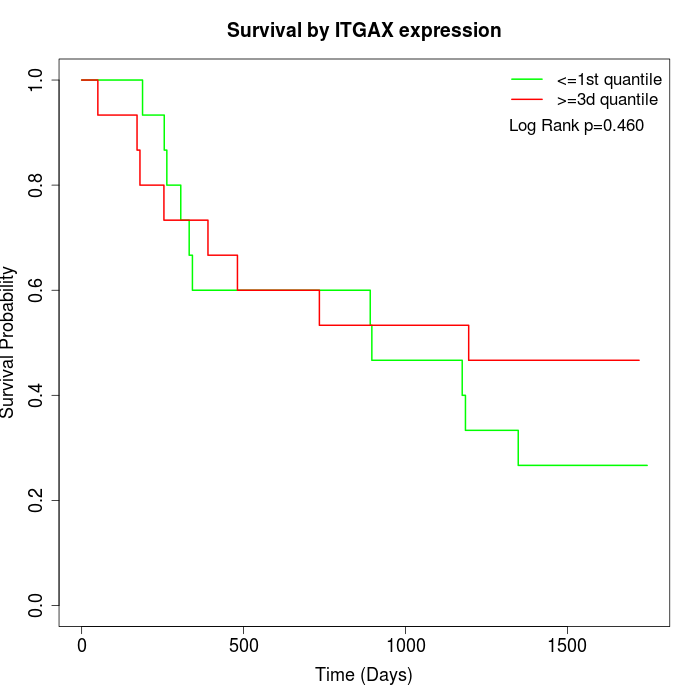
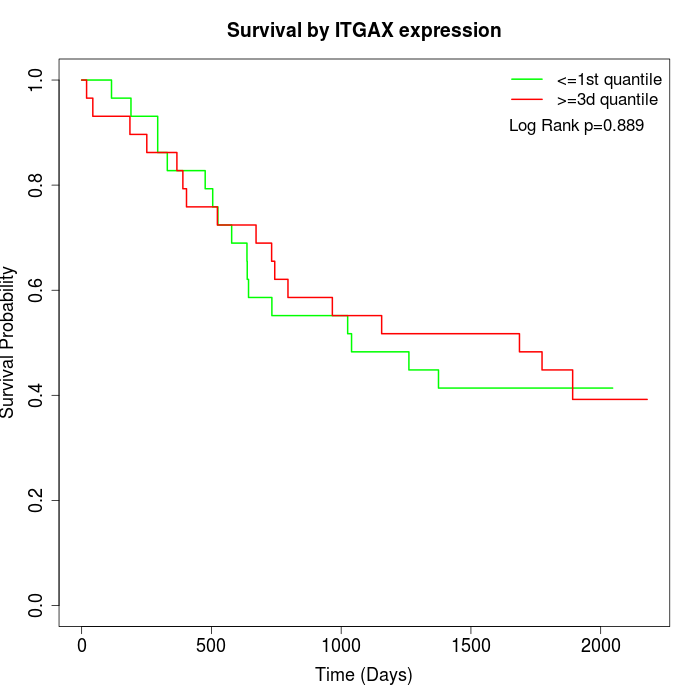
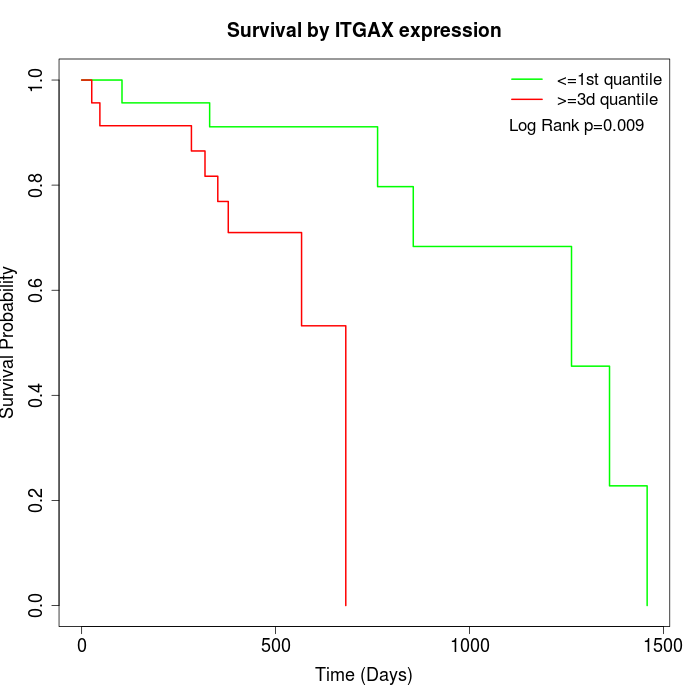
Survival by ITGAX expression:
Note: Click image to view full size file.
Copy number change of ITGAX:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ITGAX | 3687 | 4 | 5 | 21 | |
| GSE20123 | ITGAX | 3687 | 4 | 4 | 22 | |
| GSE43470 | ITGAX | 3687 | 3 | 3 | 37 | |
| GSE46452 | ITGAX | 3687 | 38 | 1 | 20 | |
| GSE47630 | ITGAX | 3687 | 11 | 7 | 22 | |
| GSE54993 | ITGAX | 3687 | 2 | 5 | 63 | |
| GSE54994 | ITGAX | 3687 | 4 | 10 | 39 | |
| GSE60625 | ITGAX | 3687 | 4 | 0 | 7 | |
| GSE74703 | ITGAX | 3687 | 3 | 2 | 31 | |
| GSE74704 | ITGAX | 3687 | 2 | 2 | 16 | |
| TCGA | ITGAX | 3687 | 20 | 10 | 66 |
Total number of gains: 95; Total number of losses: 49; Total Number of normals: 344.
Somatic mutations of ITGAX:
Generating mutation plots.
Highly correlated genes for ITGAX:
Showing top 20/416 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ITGAX | MS4A14 | 0.757527 | 3 | 0 | 3 |
| ITGAX | FERMT3 | 0.686322 | 4 | 0 | 3 |
| ITGAX | LAIR1 | 0.678898 | 6 | 0 | 5 |
| ITGAX | KIR2DL4 | 0.678181 | 3 | 0 | 3 |
| ITGAX | HK3 | 0.678088 | 8 | 0 | 7 |
| ITGAX | DVL2 | 0.657353 | 3 | 0 | 3 |
| ITGAX | CCR5 | 0.656727 | 6 | 0 | 4 |
| ITGAX | SLC7A7 | 0.646913 | 11 | 0 | 9 |
| ITGAX | PLEKHG2 | 0.643292 | 3 | 0 | 3 |
| ITGAX | RAD1 | 0.637628 | 3 | 0 | 3 |
| ITGAX | SLAMF8 | 0.634327 | 10 | 0 | 9 |
| ITGAX | SNAI1 | 0.628618 | 3 | 0 | 3 |
| ITGAX | SPATA17 | 0.627108 | 3 | 0 | 3 |
| ITGAX | COL18A1 | 0.625786 | 5 | 0 | 3 |
| ITGAX | FCER1G | 0.623731 | 9 | 0 | 8 |
| ITGAX | EFTUD2 | 0.622185 | 4 | 0 | 3 |
| ITGAX | PREX1 | 0.616551 | 4 | 0 | 3 |
| ITGAX | STIM2 | 0.616129 | 3 | 0 | 3 |
| ITGAX | FCGR3B | 0.614765 | 8 | 0 | 8 |
| ITGAX | MFSD12 | 0.613417 | 8 | 0 | 6 |
For details and further investigation, click here