| Full name: myosin heavy chain 9 | Alias Symbol: NMMHCA|NMHC-II-A|MHA|FTNS|EPSTS | ||
| Type: protein-coding gene | Cytoband: 22q12.3 | ||
| Entrez ID: 4627 | HGNC ID: HGNC:7579 | Ensembl Gene: ENSG00000100345 | OMIM ID: 160775 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
MYH9 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04530 | Tight junction | |
| hsa04810 | Regulation of actin cytoskeleton | |
| hsa05132 | Salmonella infection |
Expression of MYH9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MYH9 | 4627 | 211926_s_at | 0.5664 | 0.2281 | |
| GSE20347 | MYH9 | 4627 | 211926_s_at | 0.5833 | 0.0032 | |
| GSE23400 | MYH9 | 4627 | 211926_s_at | 0.7203 | 0.0000 | |
| GSE26886 | MYH9 | 4627 | 211926_s_at | 1.3258 | 0.0000 | |
| GSE29001 | MYH9 | 4627 | 211926_s_at | 0.5319 | 0.0334 | |
| GSE38129 | MYH9 | 4627 | 211926_s_at | 0.6880 | 0.0000 | |
| GSE45670 | MYH9 | 4627 | 211926_s_at | 0.7117 | 0.0005 | |
| GSE53622 | MYH9 | 4627 | 43265 | 0.1115 | 0.1577 | |
| GSE53624 | MYH9 | 4627 | 43265 | 0.0133 | 0.8544 | |
| GSE63941 | MYH9 | 4627 | 211926_s_at | -0.4393 | 0.5189 | |
| GSE77861 | MYH9 | 4627 | 211926_s_at | 1.2496 | 0.0066 | |
| GSE97050 | MYH9 | 4627 | A_33_P3353692 | -0.8419 | 0.2525 | |
| SRP007169 | MYH9 | 4627 | RNAseq | 3.0742 | 0.0000 | |
| SRP008496 | MYH9 | 4627 | RNAseq | 2.7024 | 0.0000 | |
| SRP064894 | MYH9 | 4627 | RNAseq | 0.9474 | 0.0001 | |
| SRP133303 | MYH9 | 4627 | RNAseq | 0.7495 | 0.0013 | |
| SRP159526 | MYH9 | 4627 | RNAseq | 0.4522 | 0.0251 | |
| SRP193095 | MYH9 | 4627 | RNAseq | 1.0202 | 0.0000 | |
| SRP219564 | MYH9 | 4627 | RNAseq | 0.5958 | 0.1534 | |
| TCGA | MYH9 | 4627 | RNAseq | 0.1148 | 0.0055 |
Upregulated datasets: 5; Downregulated datasets: 0.
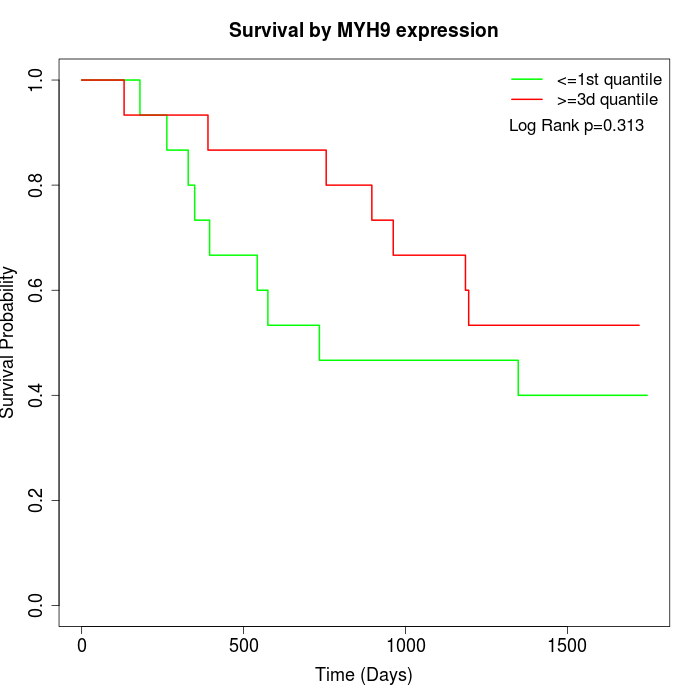
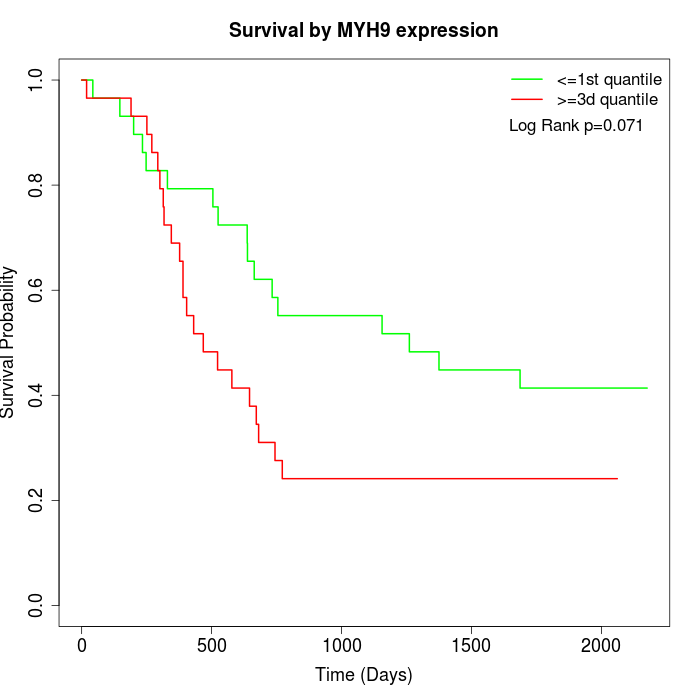
Survival by MYH9 expression:
Note: Click image to view full size file.
Copy number change of MYH9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MYH9 | 4627 | 6 | 6 | 18 | |
| GSE20123 | MYH9 | 4627 | 6 | 5 | 19 | |
| GSE43470 | MYH9 | 4627 | 4 | 6 | 33 | |
| GSE46452 | MYH9 | 4627 | 31 | 1 | 27 | |
| GSE47630 | MYH9 | 4627 | 8 | 5 | 27 | |
| GSE54993 | MYH9 | 4627 | 3 | 6 | 61 | |
| GSE54994 | MYH9 | 4627 | 9 | 9 | 35 | |
| GSE60625 | MYH9 | 4627 | 5 | 0 | 6 | |
| GSE74703 | MYH9 | 4627 | 4 | 4 | 28 | |
| GSE74704 | MYH9 | 4627 | 3 | 2 | 15 | |
| TCGA | MYH9 | 4627 | 27 | 15 | 54 |
Total number of gains: 106; Total number of losses: 59; Total Number of normals: 323.
Somatic mutations of MYH9:
Generating mutation plots.
Highly correlated genes for MYH9:
Showing top 20/807 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MYH9 | WDR54 | 0.882066 | 4 | 0 | 4 |
| MYH9 | XPR1 | 0.851945 | 4 | 0 | 4 |
| MYH9 | POLE4 | 0.796879 | 4 | 0 | 4 |
| MYH9 | SPRR2G | 0.787699 | 3 | 0 | 3 |
| MYH9 | GTF2IRD2 | 0.786311 | 3 | 0 | 3 |
| MYH9 | ALOXE3 | 0.749143 | 3 | 0 | 3 |
| MYH9 | TENM2 | 0.744233 | 4 | 0 | 4 |
| MYH9 | DLX4 | 0.740186 | 3 | 0 | 3 |
| MYH9 | YDJC | 0.735656 | 3 | 0 | 3 |
| MYH9 | NTMT1 | 0.728984 | 4 | 0 | 4 |
| MYH9 | TTYH3 | 0.721494 | 5 | 0 | 4 |
| MYH9 | GALNT18 | 0.721079 | 5 | 0 | 4 |
| MYH9 | AGO2 | 0.719876 | 3 | 0 | 3 |
| MYH9 | CKAP5 | 0.719698 | 8 | 0 | 8 |
| MYH9 | HOMER3 | 0.713195 | 10 | 0 | 9 |
| MYH9 | ITM2C | 0.705168 | 3 | 0 | 3 |
| MYH9 | BLACAT1 | 0.704512 | 4 | 0 | 4 |
| MYH9 | EMC10 | 0.704244 | 3 | 0 | 3 |
| MYH9 | SHISA2 | 0.700401 | 4 | 0 | 4 |
| MYH9 | PES1 | 0.697501 | 8 | 0 | 7 |
For details and further investigation, click here