| Full name: NK3 homeobox 2 | Alias Symbol: NKX3B|NKX3.2 | ||
| Type: protein-coding gene | Cytoband: 4p15.33 | ||
| Entrez ID: 579 | HGNC ID: HGNC:951 | Ensembl Gene: ENSG00000109705 | OMIM ID: 602183 |
| Drug and gene relationship at DGIdb | |||
Expression of NKX3-2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NKX3-2 | 579 | 207031_at | 0.0906 | 0.9229 | |
| GSE20347 | NKX3-2 | 579 | 207031_at | 0.2027 | 0.0237 | |
| GSE23400 | NKX3-2 | 579 | 207031_at | 0.0600 | 0.1545 | |
| GSE26886 | NKX3-2 | 579 | 207031_at | 0.5895 | 0.0000 | |
| GSE29001 | NKX3-2 | 579 | 207031_at | 0.2778 | 0.3329 | |
| GSE38129 | NKX3-2 | 579 | 207031_at | 0.0509 | 0.7172 | |
| GSE45670 | NKX3-2 | 579 | 207031_at | 0.0552 | 0.7030 | |
| GSE53622 | NKX3-2 | 579 | 18578 | 1.6885 | 0.0000 | |
| GSE53624 | NKX3-2 | 579 | 18578 | 1.8786 | 0.0000 | |
| GSE63941 | NKX3-2 | 579 | 207031_at | 0.3030 | 0.1972 | |
| GSE77861 | NKX3-2 | 579 | 207031_at | 0.0973 | 0.2912 | |
| SRP219564 | NKX3-2 | 579 | RNAseq | 0.3298 | 0.7652 | |
| TCGA | NKX3-2 | 579 | RNAseq | 0.9102 | 0.0239 |
Upregulated datasets: 2; Downregulated datasets: 0.
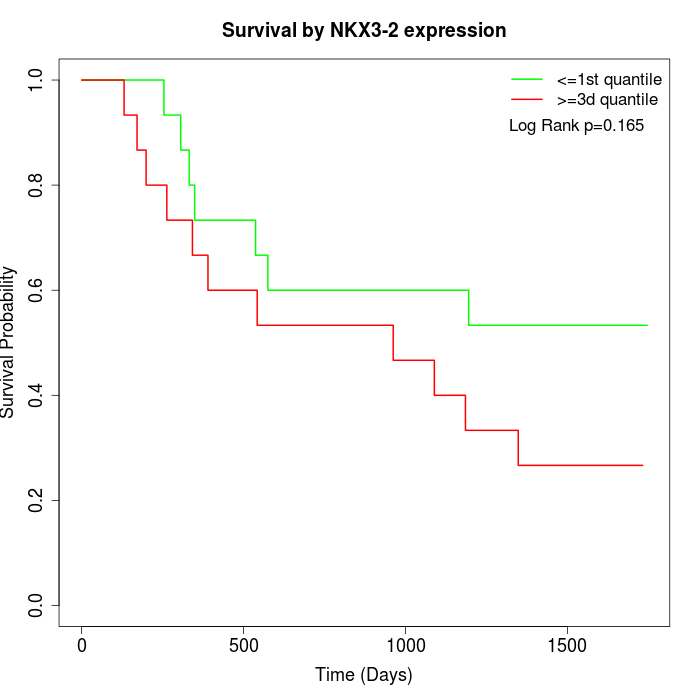
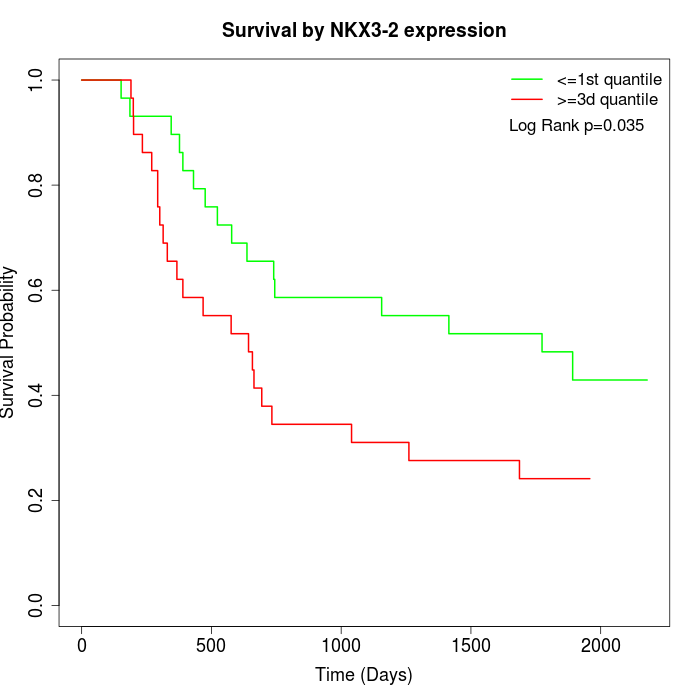
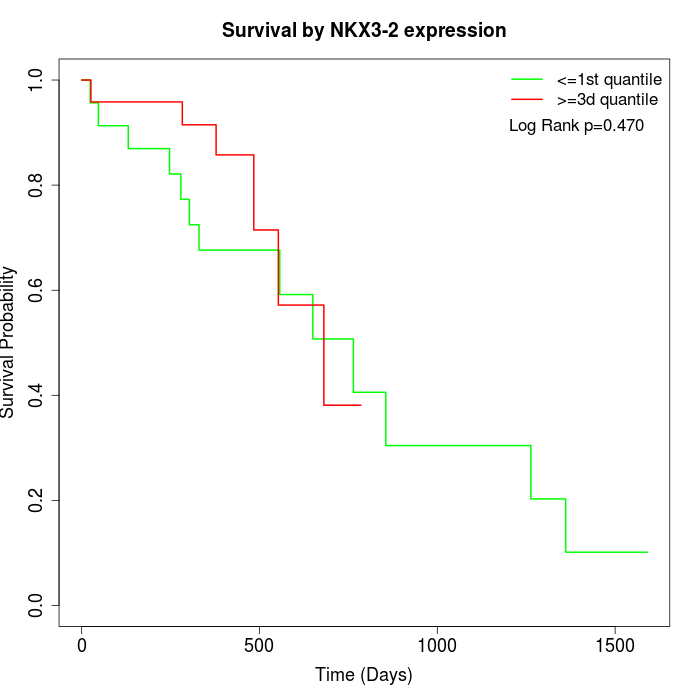
Survival by NKX3-2 expression:
Note: Click image to view full size file.
Copy number change of NKX3-2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NKX3-2 | 579 | 0 | 18 | 12 | |
| GSE20123 | NKX3-2 | 579 | 0 | 18 | 12 | |
| GSE43470 | NKX3-2 | 579 | 1 | 15 | 27 | |
| GSE46452 | NKX3-2 | 579 | 1 | 36 | 22 | |
| GSE47630 | NKX3-2 | 579 | 0 | 20 | 20 | |
| GSE54993 | NKX3-2 | 579 | 10 | 0 | 60 | |
| GSE54994 | NKX3-2 | 579 | 4 | 12 | 37 | |
| GSE60625 | NKX3-2 | 579 | 0 | 0 | 11 | |
| GSE74703 | NKX3-2 | 579 | 1 | 13 | 22 | |
| GSE74704 | NKX3-2 | 579 | 0 | 11 | 9 |
Total number of gains: 17; Total number of losses: 143; Total Number of normals: 232.
Somatic mutations of NKX3-2:
Generating mutation plots.
Highly correlated genes for NKX3-2:
Showing top 20/378 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NKX3-2 | COL10A1 | 0.716919 | 3 | 0 | 3 |
| NKX3-2 | CTHRC1 | 0.693411 | 3 | 0 | 3 |
| NKX3-2 | WIZ | 0.686774 | 3 | 0 | 3 |
| NKX3-2 | GPSM1 | 0.684152 | 3 | 0 | 3 |
| NKX3-2 | TRIM47 | 0.66096 | 3 | 0 | 3 |
| NKX3-2 | EBF4 | 0.649508 | 3 | 0 | 3 |
| NKX3-2 | B3GNT9 | 0.645173 | 5 | 0 | 5 |
| NKX3-2 | COL24A1 | 0.642753 | 4 | 0 | 4 |
| NKX3-2 | WNT9A | 0.640322 | 3 | 0 | 3 |
| NKX3-2 | CEP290 | 0.63284 | 3 | 0 | 3 |
| NKX3-2 | CLEC11A | 0.631469 | 4 | 0 | 4 |
| NKX3-2 | HTRA3 | 0.631104 | 4 | 0 | 4 |
| NKX3-2 | RHOQ | 0.627346 | 5 | 0 | 3 |
| NKX3-2 | DLX4 | 0.618604 | 6 | 0 | 5 |
| NKX3-2 | DGKG | 0.616534 | 5 | 0 | 4 |
| NKX3-2 | SCG2 | 0.614225 | 5 | 0 | 4 |
| NKX3-2 | DEDD2 | 0.613806 | 4 | 0 | 3 |
| NKX3-2 | EIF5A2 | 0.61299 | 5 | 0 | 5 |
| NKX3-2 | AMPD2 | 0.610225 | 5 | 0 | 4 |
| NKX3-2 | CXXC1 | 0.610203 | 4 | 0 | 3 |
For details and further investigation, click here