| Full name: nucleotide binding oligomerization domain containing 2 | Alias Symbol: BLAU|CD|PSORAS1|CLR16.3|NLRC2 | ||
| Type: protein-coding gene | Cytoband: 16q12.1 | ||
| Entrez ID: 64127 | HGNC ID: HGNC:5331 | Ensembl Gene: ENSG00000167207 | OMIM ID: 605956 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
NOD2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04621 | NOD-like receptor signaling pathway | |
| hsa05131 | Shigellosis | |
| hsa05152 | Tuberculosis |
Expression of NOD2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NOD2 | 64127 | 220066_at | -0.6863 | 0.6891 | |
| GSE20347 | NOD2 | 64127 | 220066_at | -1.4444 | 0.0000 | |
| GSE23400 | NOD2 | 64127 | 220066_at | -0.4888 | 0.0001 | |
| GSE26886 | NOD2 | 64127 | 220066_at | -1.6231 | 0.0004 | |
| GSE29001 | NOD2 | 64127 | 220066_at | -0.7100 | 0.0886 | |
| GSE38129 | NOD2 | 64127 | 220066_at | -0.8911 | 0.0079 | |
| GSE45670 | NOD2 | 64127 | 220066_at | 0.1949 | 0.5225 | |
| GSE53622 | NOD2 | 64127 | 102025 | -0.3301 | 0.0124 | |
| GSE53624 | NOD2 | 64127 | 10142 | -1.2338 | 0.0000 | |
| GSE63941 | NOD2 | 64127 | 220066_at | 0.4677 | 0.2653 | |
| GSE77861 | NOD2 | 64127 | 220066_at | 0.0045 | 0.9942 | |
| GSE97050 | NOD2 | 64127 | A_23_P420863 | 0.4437 | 0.5209 | |
| SRP007169 | NOD2 | 64127 | RNAseq | -2.5193 | 0.0000 | |
| SRP008496 | NOD2 | 64127 | RNAseq | -2.3426 | 0.0000 | |
| SRP064894 | NOD2 | 64127 | RNAseq | -1.1720 | 0.0001 | |
| SRP133303 | NOD2 | 64127 | RNAseq | -0.7098 | 0.0575 | |
| SRP159526 | NOD2 | 64127 | RNAseq | -0.7432 | 0.2621 | |
| SRP193095 | NOD2 | 64127 | RNAseq | -0.5815 | 0.0330 | |
| SRP219564 | NOD2 | 64127 | RNAseq | -0.5280 | 0.5543 | |
| TCGA | NOD2 | 64127 | RNAseq | 0.7273 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 6.
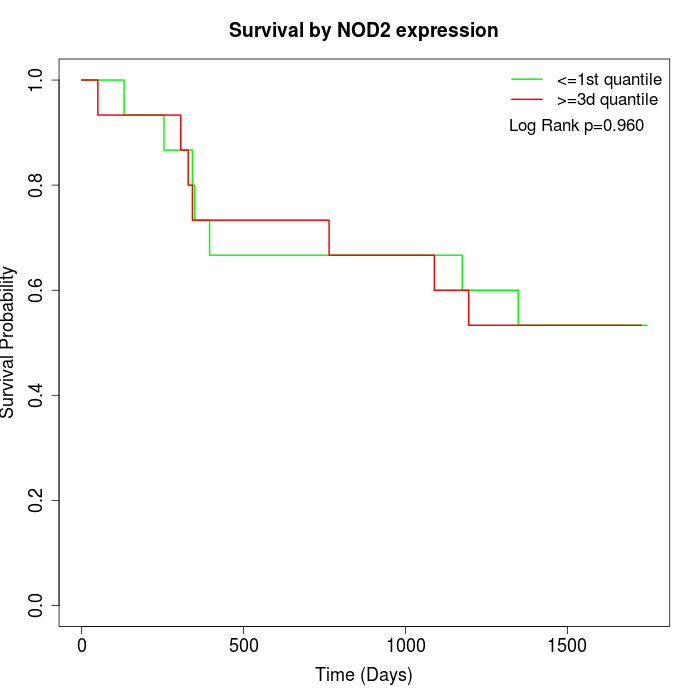
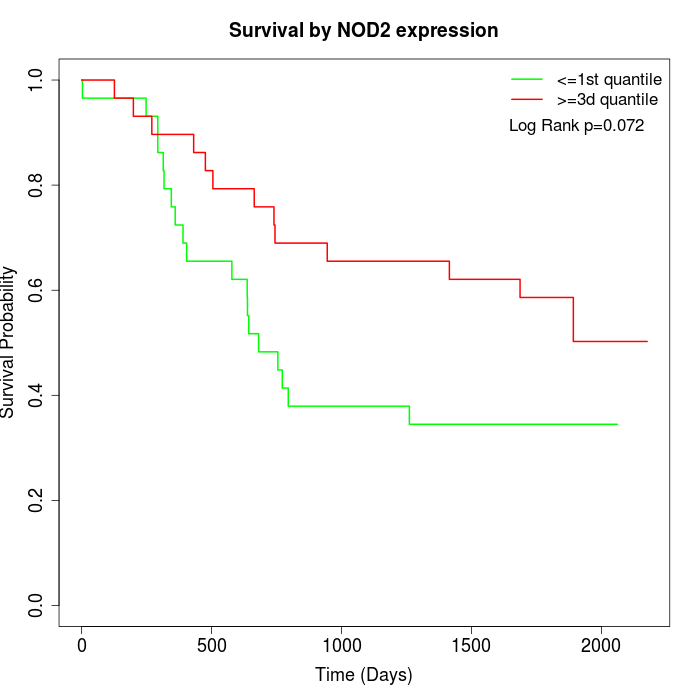
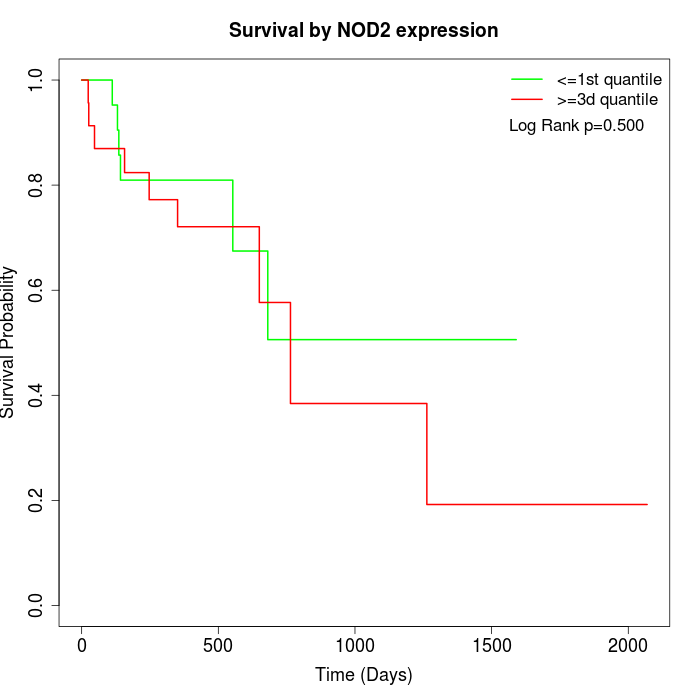
Survival by NOD2 expression:
Note: Click image to view full size file.
Copy number change of NOD2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NOD2 | 64127 | 3 | 1 | 26 | |
| GSE20123 | NOD2 | 64127 | 3 | 1 | 26 | |
| GSE43470 | NOD2 | 64127 | 2 | 7 | 34 | |
| GSE46452 | NOD2 | 64127 | 38 | 1 | 20 | |
| GSE47630 | NOD2 | 64127 | 11 | 7 | 22 | |
| GSE54993 | NOD2 | 64127 | 2 | 5 | 63 | |
| GSE54994 | NOD2 | 64127 | 6 | 10 | 37 | |
| GSE60625 | NOD2 | 64127 | 4 | 0 | 7 | |
| GSE74703 | NOD2 | 64127 | 2 | 4 | 30 | |
| GSE74704 | NOD2 | 64127 | 3 | 1 | 16 | |
| TCGA | NOD2 | 64127 | 25 | 12 | 59 |
Total number of gains: 99; Total number of losses: 49; Total Number of normals: 340.
Somatic mutations of NOD2:
Generating mutation plots.
Highly correlated genes for NOD2:
Showing top 20/461 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NOD2 | DMKN | 0.68223 | 7 | 0 | 7 |
| NOD2 | SLC10A6 | 0.677 | 3 | 0 | 3 |
| NOD2 | SLPI | 0.658471 | 12 | 0 | 10 |
| NOD2 | RNF222 | 0.657916 | 3 | 0 | 3 |
| NOD2 | SBSN | 0.657705 | 8 | 0 | 7 |
| NOD2 | C10orf99 | 0.657469 | 7 | 0 | 6 |
| NOD2 | IL36RN | 0.654269 | 11 | 0 | 10 |
| NOD2 | S100A12 | 0.652402 | 11 | 0 | 10 |
| NOD2 | SERPINB3 | 0.649537 | 8 | 0 | 6 |
| NOD2 | IL22RA1 | 0.645584 | 9 | 0 | 7 |
| NOD2 | CDA | 0.644312 | 10 | 0 | 9 |
| NOD2 | PI3 | 0.642736 | 12 | 0 | 10 |
| NOD2 | SPRR2E | 0.640152 | 4 | 0 | 4 |
| NOD2 | ALDH3B2 | 0.639941 | 11 | 0 | 10 |
| NOD2 | IL20RB | 0.638003 | 8 | 0 | 5 |
| NOD2 | SPRR2D | 0.637435 | 4 | 0 | 4 |
| NOD2 | CERS3 | 0.636068 | 4 | 0 | 4 |
| NOD2 | SPAG17 | 0.635 | 3 | 0 | 3 |
| NOD2 | IL18 | 0.633896 | 8 | 0 | 8 |
| NOD2 | SPRR2F | 0.633606 | 4 | 0 | 4 |
For details and further investigation, click here