| Full name: nucleoredoxin | Alias Symbol: FLJ12614|NRX | ||
| Type: protein-coding gene | Cytoband: 17p13.3 | ||
| Entrez ID: 64359 | HGNC ID: HGNC:18008 | Ensembl Gene: ENSG00000167693 | OMIM ID: 612895 |
| Drug and gene relationship at DGIdb | |||
Expression of NXN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | NXN | 64359 | 5995 | 0.9412 | 0.0000 | |
| GSE53624 | NXN | 64359 | 5995 | 0.8900 | 0.0000 | |
| GSE97050 | NXN | 64359 | A_23_P61778 | 0.4627 | 0.1189 | |
| SRP007169 | NXN | 64359 | RNAseq | 1.2882 | 0.0010 | |
| SRP008496 | NXN | 64359 | RNAseq | 0.7344 | 0.0461 | |
| SRP064894 | NXN | 64359 | RNAseq | 0.6964 | 0.0002 | |
| SRP133303 | NXN | 64359 | RNAseq | 0.8349 | 0.0102 | |
| SRP159526 | NXN | 64359 | RNAseq | 0.1762 | 0.6835 | |
| SRP193095 | NXN | 64359 | RNAseq | 0.7611 | 0.0050 | |
| SRP219564 | NXN | 64359 | RNAseq | 0.5454 | 0.1641 | |
| TCGA | NXN | 64359 | RNAseq | 0.1687 | 0.0239 |
Upregulated datasets: 1; Downregulated datasets: 0.
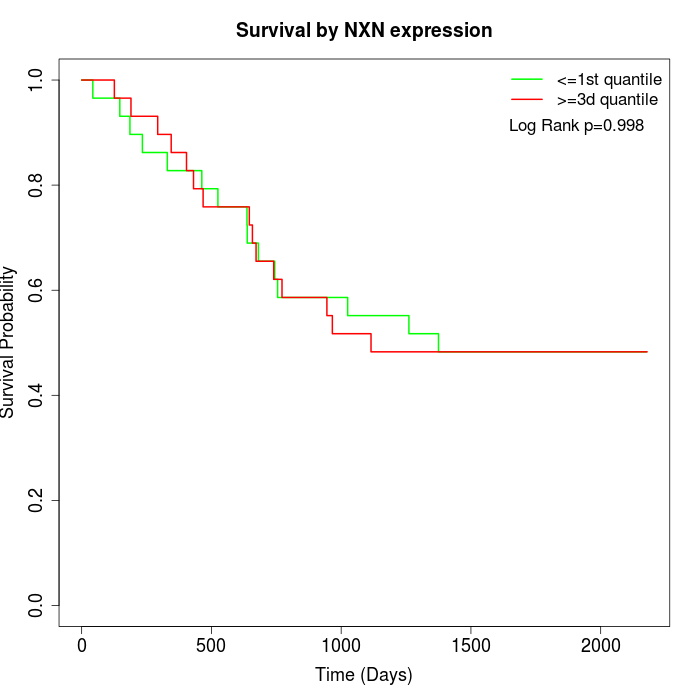
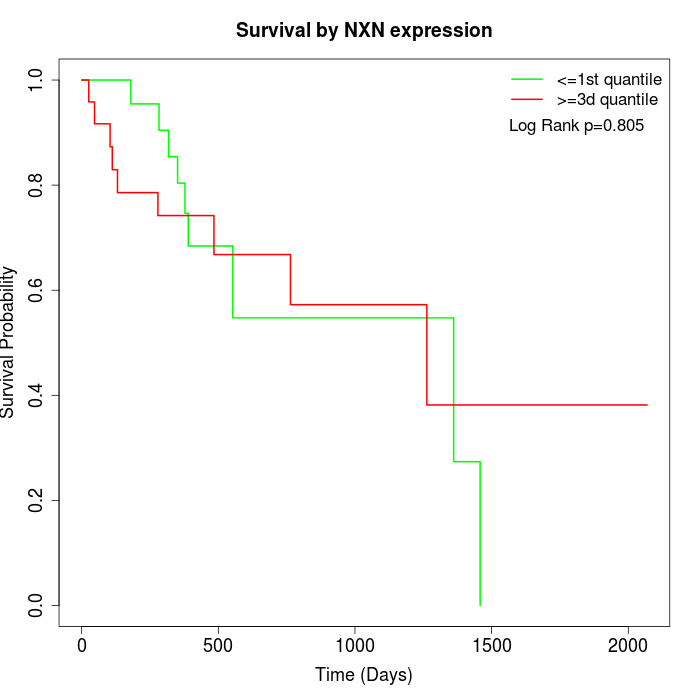
Survival by NXN expression:
Note: Click image to view full size file.
Copy number change of NXN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NXN | 64359 | 5 | 2 | 23 | |
| GSE20123 | NXN | 64359 | 5 | 3 | 22 | |
| GSE43470 | NXN | 64359 | 3 | 7 | 33 | |
| GSE46452 | NXN | 64359 | 34 | 1 | 24 | |
| GSE47630 | NXN | 64359 | 8 | 1 | 31 | |
| GSE54993 | NXN | 64359 | 8 | 3 | 59 | |
| GSE54994 | NXN | 64359 | 7 | 9 | 37 | |
| GSE60625 | NXN | 64359 | 4 | 0 | 7 | |
| GSE74703 | NXN | 64359 | 2 | 4 | 30 | |
| GSE74704 | NXN | 64359 | 3 | 1 | 16 | |
| TCGA | NXN | 64359 | 19 | 21 | 56 |
Total number of gains: 98; Total number of losses: 52; Total Number of normals: 338.
Somatic mutations of NXN:
Generating mutation plots.
Highly correlated genes for NXN:
Showing top 20/160 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NXN | MSN | 0.796913 | 3 | 0 | 3 |
| NXN | PANX1 | 0.790106 | 3 | 0 | 3 |
| NXN | CALU | 0.769804 | 3 | 0 | 3 |
| NXN | RIN2 | 0.761506 | 3 | 0 | 3 |
| NXN | STK3 | 0.75745 | 3 | 0 | 3 |
| NXN | PTGFRN | 0.756047 | 3 | 0 | 3 |
| NXN | B3GNT9 | 0.751654 | 3 | 0 | 3 |
| NXN | TRIO | 0.749911 | 3 | 0 | 3 |
| NXN | ACLY | 0.744429 | 3 | 0 | 3 |
| NXN | LPCAT1 | 0.731345 | 3 | 0 | 3 |
| NXN | KLF7 | 0.729335 | 3 | 0 | 3 |
| NXN | PTK2 | 0.725862 | 3 | 0 | 3 |
| NXN | CDK6 | 0.723711 | 3 | 0 | 3 |
| NXN | ARSI | 0.723685 | 3 | 0 | 3 |
| NXN | FNDC3B | 0.721491 | 3 | 0 | 3 |
| NXN | ANGPTL2 | 0.71996 | 3 | 0 | 3 |
| NXN | ASAP1 | 0.719568 | 3 | 0 | 3 |
| NXN | SPARC | 0.716997 | 3 | 0 | 3 |
| NXN | COL5A1 | 0.71307 | 3 | 0 | 3 |
| NXN | AGRN | 0.710864 | 3 | 0 | 3 |
For details and further investigation, click here