| Full name: phosphodiesterase 4D | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 5q11.2-q12.1 | ||
| Entrez ID: 5144 | HGNC ID: HGNC:8783 | Ensembl Gene: ENSG00000113448 | OMIM ID: 600129 |
| Drug and gene relationship at DGIdb | |||
PDE4D involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04024 | cAMP signaling pathway |
Expression of PDE4D:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PDE4D | 5144 | 204491_at | -0.5714 | 0.2077 | |
| GSE20347 | PDE4D | 5144 | 204491_at | -0.1125 | 0.5466 | |
| GSE23400 | PDE4D | 5144 | 204491_at | -0.0556 | 0.4961 | |
| GSE26886 | PDE4D | 5144 | 204491_at | 0.9734 | 0.0000 | |
| GSE29001 | PDE4D | 5144 | 204491_at | 0.1754 | 0.3810 | |
| GSE38129 | PDE4D | 5144 | 204491_at | -0.3614 | 0.0943 | |
| GSE45670 | PDE4D | 5144 | 204491_at | -0.8267 | 0.0042 | |
| GSE53622 | PDE4D | 5144 | 127256 | -0.8164 | 0.0000 | |
| GSE53624 | PDE4D | 5144 | 67237 | -0.3613 | 0.0000 | |
| GSE63941 | PDE4D | 5144 | 204491_at | -1.8564 | 0.0304 | |
| GSE77861 | PDE4D | 5144 | 204491_at | 0.4829 | 0.0201 | |
| GSE97050 | PDE4D | 5144 | A_33_P3389649 | 0.5317 | 0.2252 | |
| SRP007169 | PDE4D | 5144 | RNAseq | 0.6163 | 0.3070 | |
| SRP008496 | PDE4D | 5144 | RNAseq | 0.6607 | 0.0939 | |
| SRP064894 | PDE4D | 5144 | RNAseq | -0.0609 | 0.8035 | |
| SRP133303 | PDE4D | 5144 | RNAseq | -0.1686 | 0.4611 | |
| SRP159526 | PDE4D | 5144 | RNAseq | -0.2114 | 0.6665 | |
| SRP193095 | PDE4D | 5144 | RNAseq | -0.1371 | 0.5155 | |
| SRP219564 | PDE4D | 5144 | RNAseq | 1.9417 | 0.0043 | |
| TCGA | PDE4D | 5144 | RNAseq | -0.5128 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 1.
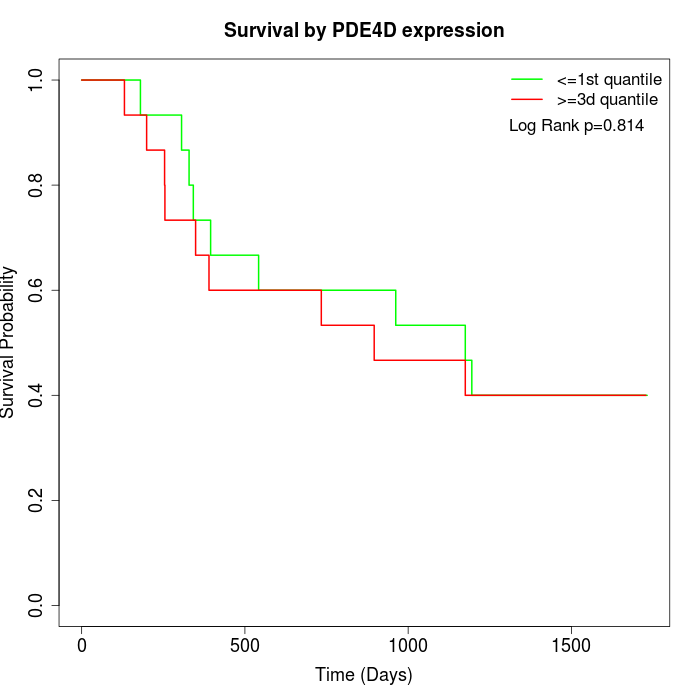
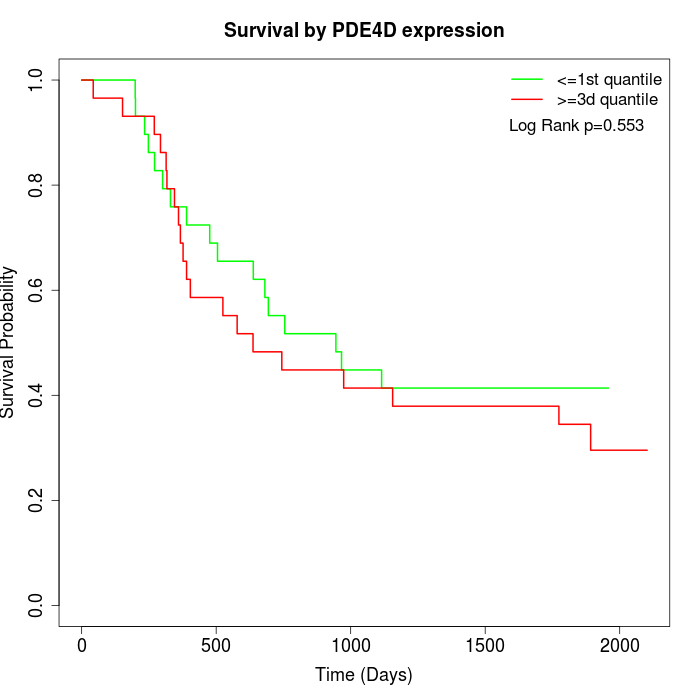
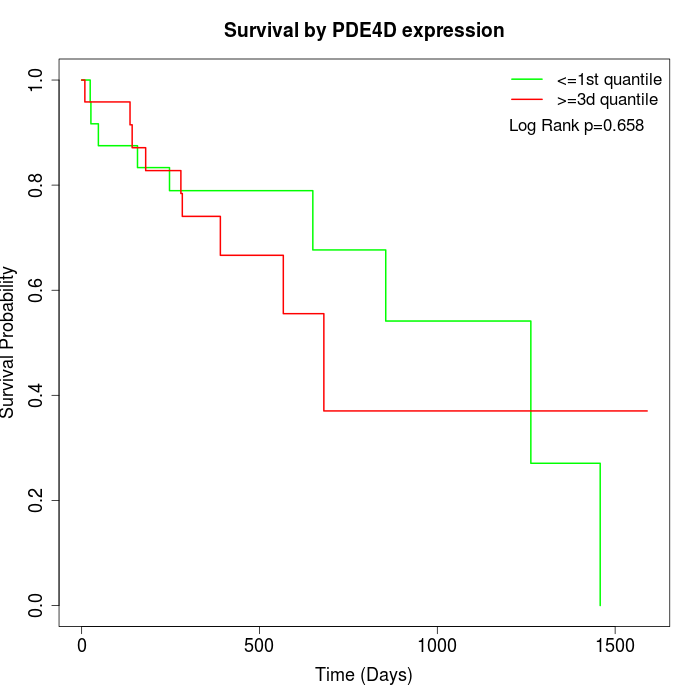
Survival by PDE4D expression:
Note: Click image to view full size file.
Copy number change of PDE4D:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PDE4D | 5144 | 0 | 13 | 17 | |
| GSE20123 | PDE4D | 5144 | 0 | 13 | 17 | |
| GSE43470 | PDE4D | 5144 | 1 | 12 | 30 | |
| GSE46452 | PDE4D | 5144 | 0 | 28 | 31 | |
| GSE47630 | PDE4D | 5144 | 2 | 17 | 21 | |
| GSE54993 | PDE4D | 5144 | 8 | 1 | 61 | |
| GSE54994 | PDE4D | 5144 | 2 | 19 | 32 | |
| GSE60625 | PDE4D | 5144 | 0 | 0 | 11 | |
| GSE74703 | PDE4D | 5144 | 1 | 9 | 26 | |
| GSE74704 | PDE4D | 5144 | 0 | 6 | 14 | |
| TCGA | PDE4D | 5144 | 6 | 52 | 38 |
Total number of gains: 20; Total number of losses: 170; Total Number of normals: 298.
Somatic mutations of PDE4D:
Generating mutation plots.
Highly correlated genes for PDE4D:
Showing top 20/324 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PDE4D | MSRB3 | 0.711317 | 3 | 0 | 3 |
| PDE4D | C3orf70 | 0.703875 | 4 | 0 | 4 |
| PDE4D | FAXC | 0.693618 | 5 | 0 | 5 |
| PDE4D | NR2F1-AS1 | 0.6909 | 5 | 0 | 5 |
| PDE4D | HSPB6 | 0.686334 | 4 | 0 | 4 |
| PDE4D | GSTA3 | 0.676987 | 3 | 0 | 3 |
| PDE4D | KCNS3 | 0.676399 | 3 | 0 | 3 |
| PDE4D | PDLIM3 | 0.674681 | 8 | 0 | 8 |
| PDE4D | HSPB7 | 0.674405 | 6 | 0 | 5 |
| PDE4D | KRT8 | 0.671554 | 3 | 0 | 3 |
| PDE4D | NIPA2 | 0.669703 | 3 | 0 | 3 |
| PDE4D | KLHL41 | 0.667091 | 3 | 0 | 3 |
| PDE4D | PARVA | 0.666159 | 8 | 0 | 7 |
| PDE4D | LPP | 0.663833 | 8 | 0 | 7 |
| PDE4D | PNMA2 | 0.663142 | 4 | 0 | 3 |
| PDE4D | AFF2 | 0.661643 | 3 | 0 | 3 |
| PDE4D | MAP1B | 0.653763 | 8 | 0 | 7 |
| PDE4D | FHL1 | 0.648288 | 7 | 0 | 6 |
| PDE4D | DIXDC1 | 0.648135 | 7 | 0 | 6 |
| PDE4D | AOC3 | 0.647621 | 7 | 0 | 7 |
For details and further investigation, click here