| Full name: peptidylprolyl isomerase F | Alias Symbol: hCyP3|Cyp-D | ||
| Type: protein-coding gene | Cytoband: 10q22.3 | ||
| Entrez ID: 10105 | HGNC ID: HGNC:9259 | Ensembl Gene: ENSG00000108179 | OMIM ID: 604486 |
| Drug and gene relationship at DGIdb | |||
PPIF involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04022 | cGMP-PKG signaling pathway | |
| hsa05012 | Parkinson's disease | |
| hsa05145 | Toxoplasmosis |
Expression of PPIF:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PPIF | 10105 | 201489_at | 1.1338 | 0.0937 | |
| GSE20347 | PPIF | 10105 | 201489_at | 0.5446 | 0.0310 | |
| GSE23400 | PPIF | 10105 | 201489_at | 0.2234 | 0.0814 | |
| GSE26886 | PPIF | 10105 | 201489_at | -0.6881 | 0.0885 | |
| GSE29001 | PPIF | 10105 | 201489_at | -0.2549 | 0.4132 | |
| GSE38129 | PPIF | 10105 | 201489_at | 0.7624 | 0.0003 | |
| GSE45670 | PPIF | 10105 | 201489_at | 1.0216 | 0.0092 | |
| GSE53622 | PPIF | 10105 | 26130 | 0.2689 | 0.0809 | |
| GSE53624 | PPIF | 10105 | 26130 | 0.1980 | 0.0612 | |
| GSE63941 | PPIF | 10105 | 201489_at | 0.6370 | 0.1290 | |
| GSE77861 | PPIF | 10105 | 201489_at | 0.2789 | 0.3382 | |
| GSE97050 | PPIF | 10105 | A_23_P202104 | 0.3657 | 0.2809 | |
| SRP007169 | PPIF | 10105 | RNAseq | -2.8512 | 0.0000 | |
| SRP008496 | PPIF | 10105 | RNAseq | -2.3904 | 0.0000 | |
| SRP064894 | PPIF | 10105 | RNAseq | -0.6966 | 0.0110 | |
| SRP133303 | PPIF | 10105 | RNAseq | 0.5666 | 0.0407 | |
| SRP159526 | PPIF | 10105 | RNAseq | 0.4154 | 0.4666 | |
| SRP193095 | PPIF | 10105 | RNAseq | -0.4046 | 0.0558 | |
| SRP219564 | PPIF | 10105 | RNAseq | -0.1386 | 0.8299 | |
| TCGA | PPIF | 10105 | RNAseq | 0.0570 | 0.4124 |
Upregulated datasets: 1; Downregulated datasets: 2.
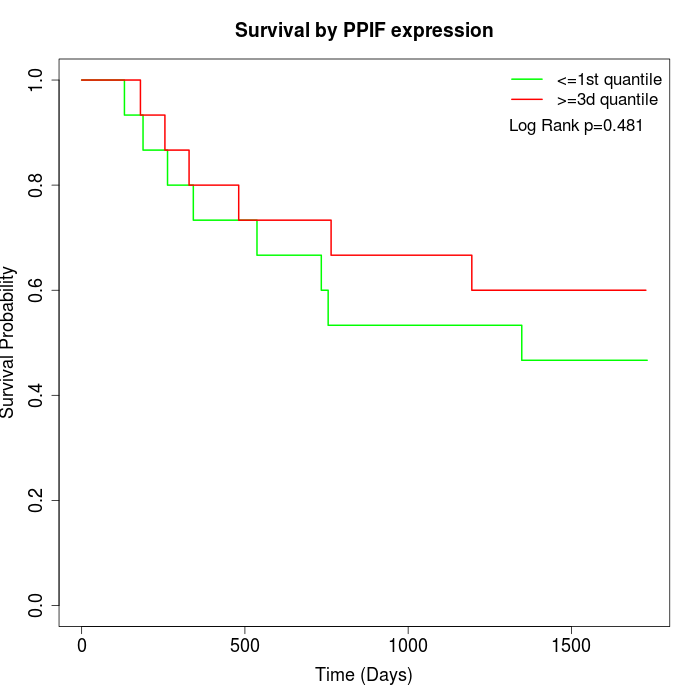
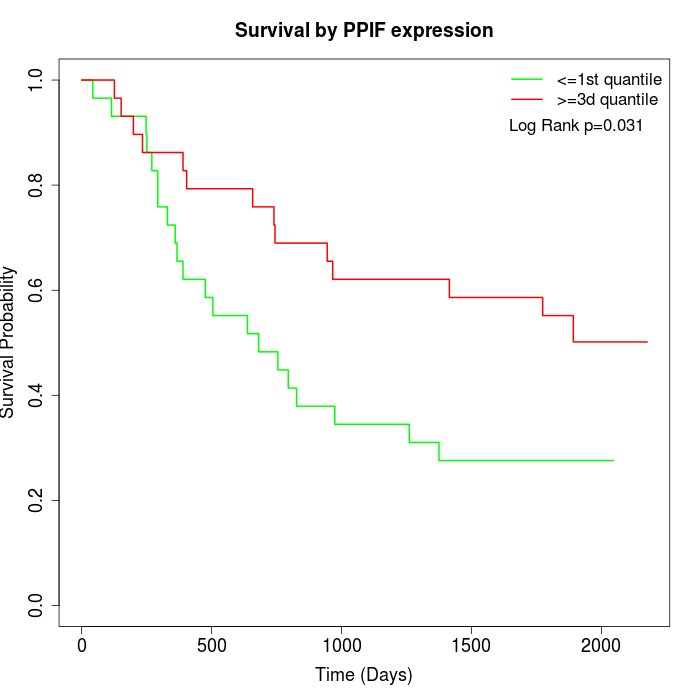
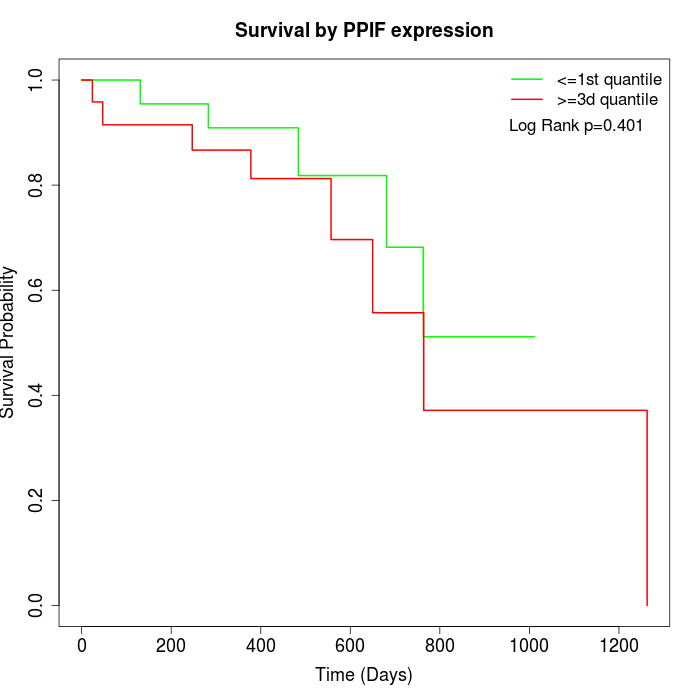
Survival by PPIF expression:
Note: Click image to view full size file.
Copy number change of PPIF:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PPIF | 10105 | 2 | 5 | 23 | |
| GSE20123 | PPIF | 10105 | 2 | 4 | 24 | |
| GSE43470 | PPIF | 10105 | 1 | 7 | 35 | |
| GSE46452 | PPIF | 10105 | 0 | 11 | 48 | |
| GSE47630 | PPIF | 10105 | 2 | 14 | 24 | |
| GSE54993 | PPIF | 10105 | 7 | 0 | 63 | |
| GSE54994 | PPIF | 10105 | 1 | 11 | 41 | |
| GSE60625 | PPIF | 10105 | 0 | 0 | 11 | |
| GSE74703 | PPIF | 10105 | 1 | 4 | 31 | |
| GSE74704 | PPIF | 10105 | 1 | 2 | 17 | |
| TCGA | PPIF | 10105 | 10 | 24 | 62 |
Total number of gains: 27; Total number of losses: 82; Total Number of normals: 379.
Somatic mutations of PPIF:
Generating mutation plots.
Highly correlated genes for PPIF:
Showing top 20/320 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PPIF | REL | 0.733864 | 3 | 0 | 3 |
| PPIF | TM2D2 | 0.717414 | 3 | 0 | 3 |
| PPIF | NT5E | 0.687773 | 3 | 0 | 3 |
| PPIF | MCTP2 | 0.674626 | 3 | 0 | 3 |
| PPIF | LRIF1 | 0.674446 | 3 | 0 | 3 |
| PPIF | SLC7A6 | 0.657402 | 3 | 0 | 3 |
| PPIF | RFWD3 | 0.65661 | 6 | 0 | 6 |
| PPIF | AGTRAP | 0.650371 | 3 | 0 | 3 |
| PPIF | CCDC86 | 0.648943 | 6 | 0 | 6 |
| PPIF | ADAMTS12 | 0.645245 | 3 | 0 | 3 |
| PPIF | CBFB | 0.644223 | 4 | 0 | 3 |
| PPIF | PSMG3 | 0.638447 | 4 | 0 | 4 |
| PPIF | ZBTB9 | 0.629198 | 4 | 0 | 3 |
| PPIF | NUS1 | 0.626193 | 3 | 0 | 3 |
| PPIF | DNAJB11 | 0.623135 | 3 | 0 | 3 |
| PPIF | LRRC59 | 0.619467 | 6 | 0 | 5 |
| PPIF | VAC14 | 0.610968 | 5 | 0 | 4 |
| PPIF | LIPG | 0.609859 | 5 | 0 | 4 |
| PPIF | HMGA2 | 0.604775 | 5 | 0 | 4 |
| PPIF | ADRM1 | 0.604563 | 8 | 0 | 7 |
For details and further investigation, click here