| Full name: RAS guanyl releasing protein 3 | Alias Symbol: KIAA0846|GRP3|CalDAG-GEFIII | ||
| Type: protein-coding gene | Cytoband: 2p22.3 | ||
| Entrez ID: 25780 | HGNC ID: HGNC:14545 | Ensembl Gene: ENSG00000152689 | OMIM ID: 609531 |
| Drug and gene relationship at DGIdb | |||
RASGRP3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04014 | Ras signaling pathway | |
| hsa04015 | Rap1 signaling pathway | |
| hsa04662 | B cell receptor signaling pathway |
Expression of RASGRP3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RASGRP3 | 25780 | 205801_s_at | 0.4624 | 0.3411 | |
| GSE20347 | RASGRP3 | 25780 | 205801_s_at | 0.0579 | 0.5059 | |
| GSE23400 | RASGRP3 | 25780 | 205801_s_at | 0.0737 | 0.0323 | |
| GSE26886 | RASGRP3 | 25780 | 205801_s_at | -0.5879 | 0.0119 | |
| GSE29001 | RASGRP3 | 25780 | 205801_s_at | 0.4178 | 0.1337 | |
| GSE38129 | RASGRP3 | 25780 | 205801_s_at | 0.0022 | 0.9909 | |
| GSE45670 | RASGRP3 | 25780 | 205801_s_at | -0.1117 | 0.6915 | |
| GSE53622 | RASGRP3 | 25780 | 34290 | 0.2379 | 0.0822 | |
| GSE53624 | RASGRP3 | 25780 | 34290 | 0.1719 | 0.2649 | |
| GSE63941 | RASGRP3 | 25780 | 244526_at | 0.1760 | 0.2231 | |
| GSE77861 | RASGRP3 | 25780 | 244526_at | -0.1684 | 0.2967 | |
| GSE97050 | RASGRP3 | 25780 | A_33_P3365582 | 0.3777 | 0.2396 | |
| SRP007169 | RASGRP3 | 25780 | RNAseq | 2.4620 | 0.0002 | |
| SRP008496 | RASGRP3 | 25780 | RNAseq | 1.8886 | 0.0001 | |
| SRP064894 | RASGRP3 | 25780 | RNAseq | 1.4288 | 0.0000 | |
| SRP133303 | RASGRP3 | 25780 | RNAseq | 0.1793 | 0.4539 | |
| SRP159526 | RASGRP3 | 25780 | RNAseq | -0.0204 | 0.9560 | |
| SRP193095 | RASGRP3 | 25780 | RNAseq | 0.2141 | 0.4251 | |
| SRP219564 | RASGRP3 | 25780 | RNAseq | 0.8965 | 0.0100 | |
| TCGA | RASGRP3 | 25780 | RNAseq | -0.3801 | 0.0025 |
Upregulated datasets: 3; Downregulated datasets: 0.
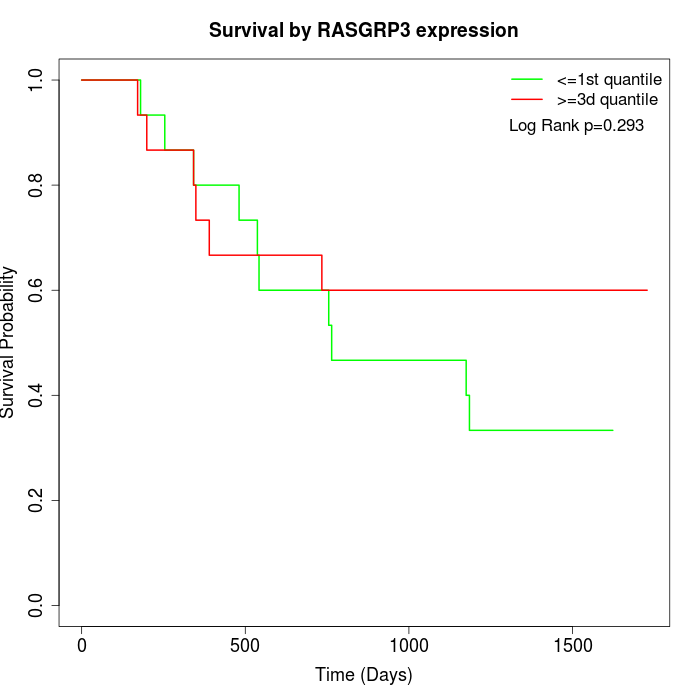
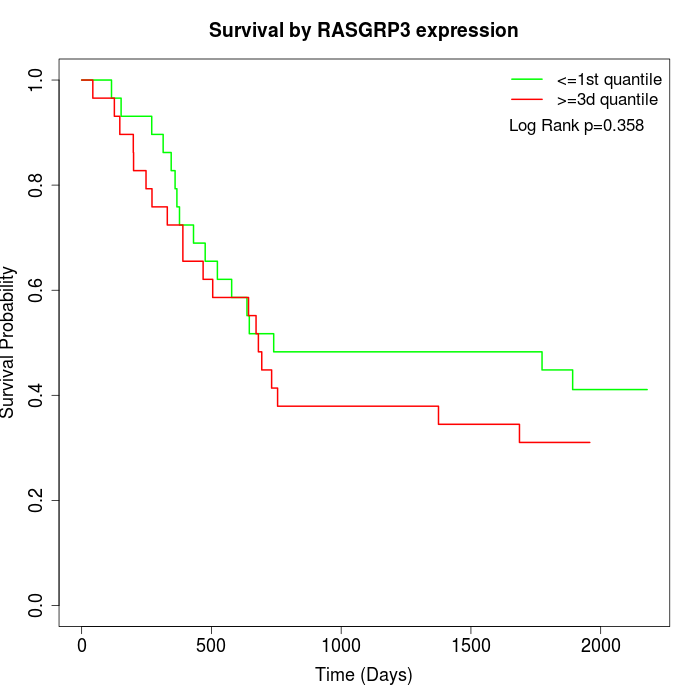
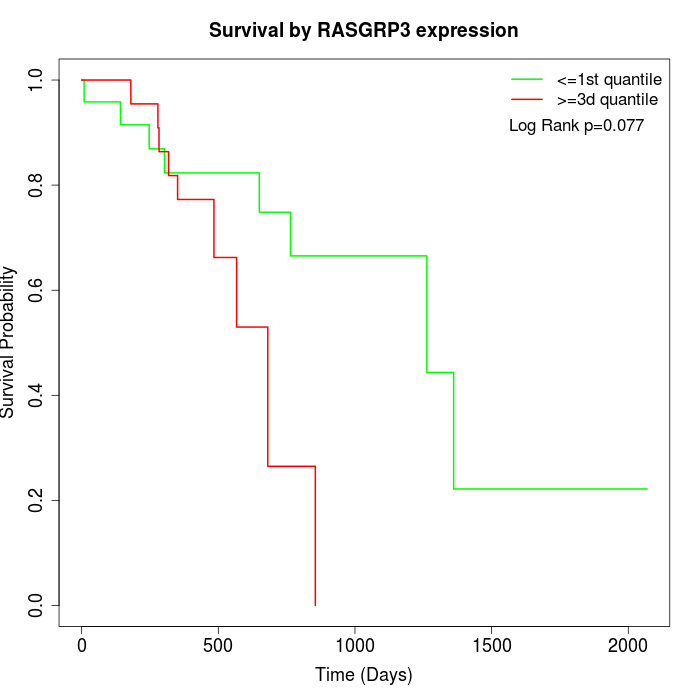
Survival by RASGRP3 expression:
Note: Click image to view full size file.
Copy number change of RASGRP3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RASGRP3 | 25780 | 10 | 1 | 19 | |
| GSE20123 | RASGRP3 | 25780 | 9 | 1 | 20 | |
| GSE43470 | RASGRP3 | 25780 | 3 | 1 | 39 | |
| GSE46452 | RASGRP3 | 25780 | 2 | 4 | 53 | |
| GSE47630 | RASGRP3 | 25780 | 7 | 0 | 33 | |
| GSE54993 | RASGRP3 | 25780 | 0 | 5 | 65 | |
| GSE54994 | RASGRP3 | 25780 | 10 | 0 | 43 | |
| GSE60625 | RASGRP3 | 25780 | 0 | 3 | 8 | |
| GSE74703 | RASGRP3 | 25780 | 3 | 0 | 33 | |
| GSE74704 | RASGRP3 | 25780 | 9 | 1 | 10 | |
| TCGA | RASGRP3 | 25780 | 36 | 3 | 57 |
Total number of gains: 89; Total number of losses: 19; Total Number of normals: 380.
Somatic mutations of RASGRP3:
Generating mutation plots.
Highly correlated genes for RASGRP3:
Showing top 20/445 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RASGRP3 | SNX20 | 0.768374 | 6 | 0 | 6 |
| RASGRP3 | C19orf38 | 0.75697 | 3 | 0 | 3 |
| RASGRP3 | CERKL | 0.753211 | 3 | 0 | 3 |
| RASGRP3 | PTPRE | 0.751248 | 3 | 0 | 3 |
| RASGRP3 | TNFAIP8L2 | 0.744726 | 6 | 0 | 6 |
| RASGRP3 | PREX1 | 0.728882 | 4 | 0 | 4 |
| RASGRP3 | ARHGAP15 | 0.71499 | 4 | 0 | 3 |
| RASGRP3 | CLEC14A | 0.714466 | 4 | 0 | 4 |
| RASGRP3 | MOGS | 0.710821 | 3 | 0 | 3 |
| RASGRP3 | GIMAP5 | 0.709978 | 4 | 0 | 3 |
| RASGRP3 | ST8SIA4 | 0.707598 | 8 | 0 | 6 |
| RASGRP3 | ARHGAP18 | 0.704412 | 4 | 0 | 4 |
| RASGRP3 | EBAG9 | 0.703425 | 3 | 0 | 3 |
| RASGRP3 | HVCN1 | 0.693856 | 4 | 0 | 3 |
| RASGRP3 | NCKAP1L | 0.693566 | 9 | 0 | 8 |
| RASGRP3 | IL10RA | 0.693516 | 9 | 0 | 9 |
| RASGRP3 | GPR141 | 0.690327 | 3 | 0 | 3 |
| RASGRP3 | FLOT2 | 0.689662 | 3 | 0 | 3 |
| RASGRP3 | RGS18 | 0.688671 | 6 | 0 | 5 |
| RASGRP3 | SH2D3C | 0.688517 | 7 | 0 | 5 |
For details and further investigation, click here