| Full name: RAD52 motif containing 1 | Alias Symbol: MGC33977 | ||
| Type: protein-coding gene | Cytoband: 17q12 | ||
| Entrez ID: 201299 | HGNC ID: HGNC:19950 | Ensembl Gene: ENSG00000278023 | OMIM ID: 612896 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of RDM1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RDM1 | 201299 | 239169_at | 0.0339 | 0.9144 | |
| GSE26886 | RDM1 | 201299 | 239169_at | -0.1822 | 0.1249 | |
| GSE45670 | RDM1 | 201299 | 239169_at | 0.3159 | 0.0182 | |
| GSE53622 | RDM1 | 201299 | 655 | 0.6699 | 0.0000 | |
| GSE53624 | RDM1 | 201299 | 655 | 0.7115 | 0.0000 | |
| GSE63941 | RDM1 | 201299 | 239169_at | 1.4525 | 0.0483 | |
| GSE77861 | RDM1 | 201299 | 239169_at | -0.0833 | 0.3616 | |
| GSE97050 | RDM1 | 201299 | A_23_P141447 | 0.0999 | 0.6797 | |
| SRP133303 | RDM1 | 201299 | RNAseq | -0.0198 | 0.9379 | |
| SRP159526 | RDM1 | 201299 | RNAseq | 1.1380 | 0.0487 | |
| SRP193095 | RDM1 | 201299 | RNAseq | -0.2543 | 0.1460 | |
| SRP219564 | RDM1 | 201299 | RNAseq | -0.0850 | 0.8399 | |
| TCGA | RDM1 | 201299 | RNAseq | 2.6699 | 0.0000 |
Upregulated datasets: 3; Downregulated datasets: 0.
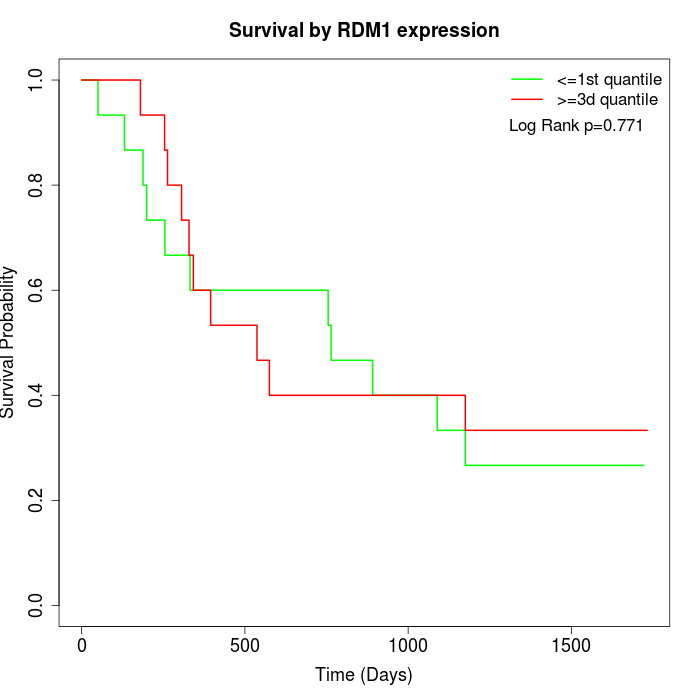
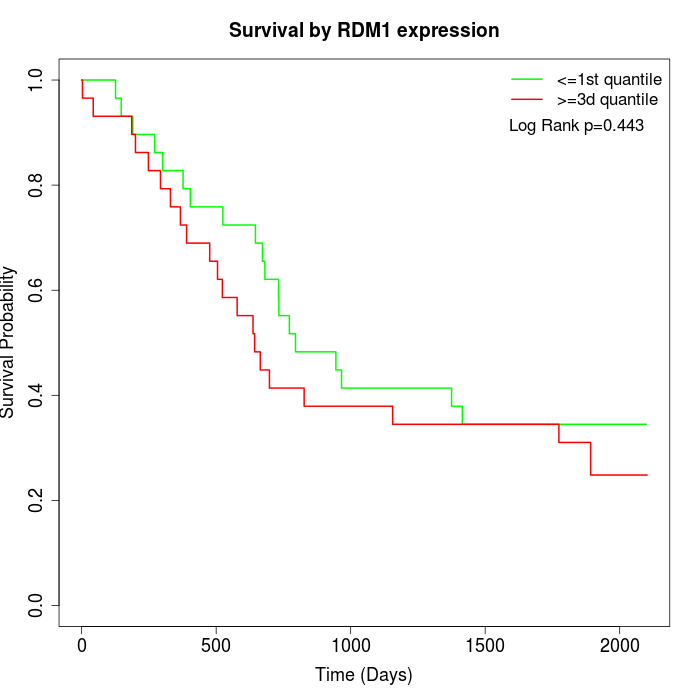
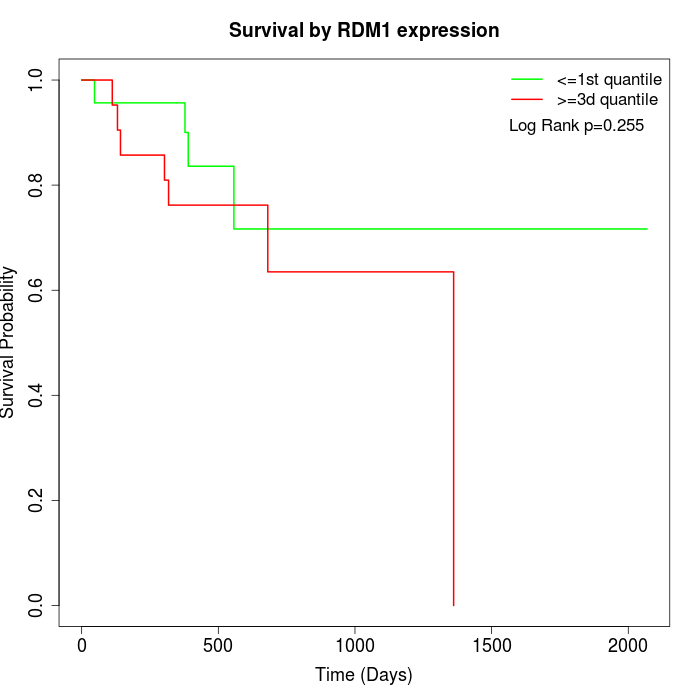
Survival by RDM1 expression:
Note: Click image to view full size file.
Copy number change of RDM1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RDM1 | 201299 | 5 | 1 | 24 | |
| GSE20123 | RDM1 | 201299 | 5 | 1 | 24 | |
| GSE43470 | RDM1 | 201299 | 1 | 2 | 40 | |
| GSE46452 | RDM1 | 201299 | 34 | 0 | 25 | |
| GSE47630 | RDM1 | 201299 | 7 | 1 | 32 | |
| GSE54993 | RDM1 | 201299 | 3 | 3 | 64 | |
| GSE54994 | RDM1 | 201299 | 8 | 6 | 39 | |
| GSE60625 | RDM1 | 201299 | 4 | 0 | 7 | |
| GSE74703 | RDM1 | 201299 | 1 | 1 | 34 | |
| GSE74704 | RDM1 | 201299 | 3 | 1 | 16 | |
| TCGA | RDM1 | 201299 | 20 | 9 | 67 |
Total number of gains: 91; Total number of losses: 25; Total Number of normals: 372.
Somatic mutations of RDM1:
Generating mutation plots.
Highly correlated genes for RDM1:
Showing top 20/30 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RDM1 | PRIM1 | 0.613007 | 3 | 0 | 3 |
| RDM1 | ASCL3 | 0.603436 | 3 | 0 | 3 |
| RDM1 | CASP12 | 0.600276 | 3 | 0 | 3 |
| RDM1 | RNPEP | 0.599467 | 3 | 0 | 3 |
| RDM1 | PRSS16 | 0.589371 | 5 | 0 | 3 |
| RDM1 | CDCA8 | 0.565904 | 5 | 0 | 4 |
| RDM1 | TMEM106C | 0.553618 | 4 | 0 | 3 |
| RDM1 | RMI2 | 0.547326 | 4 | 0 | 3 |
| RDM1 | FXYD3 | 0.541638 | 4 | 0 | 3 |
| RDM1 | CCNF | 0.538335 | 4 | 0 | 3 |
| RDM1 | GINS2 | 0.537317 | 5 | 0 | 3 |
| RDM1 | FANCA | 0.536824 | 4 | 0 | 3 |
| RDM1 | ZNF789 | 0.534724 | 5 | 0 | 3 |
| RDM1 | POLA2 | 0.53394 | 4 | 0 | 3 |
| RDM1 | MEGF10 | 0.533561 | 4 | 0 | 3 |
| RDM1 | KIF18B | 0.532932 | 4 | 0 | 3 |
| RDM1 | C5orf34 | 0.529663 | 4 | 0 | 3 |
| RDM1 | EXO1 | 0.528201 | 5 | 0 | 3 |
| RDM1 | KIF15 | 0.52759 | 4 | 0 | 3 |
| RDM1 | PAGR1 | 0.525712 | 3 | 0 | 3 |
For details and further investigation, click here