| Full name: regenerating family member 1 beta | Alias Symbol: REGL|PSPS2|REGH|REGI-BETA | ||
| Type: protein-coding gene | Cytoband: 2p12 | ||
| Entrez ID: 5968 | HGNC ID: HGNC:9952 | Ensembl Gene: ENSG00000172023 | OMIM ID: 167771 |
| Drug and gene relationship at DGIdb | |||
Expression of REG1B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | REG1B | 5968 | 205886_at | 0.2437 | 0.2874 | |
| GSE20347 | REG1B | 5968 | 205886_at | 0.1006 | 0.1489 | |
| GSE23400 | REG1B | 5968 | 205886_at | -0.0325 | 0.5155 | |
| GSE26886 | REG1B | 5968 | 205886_at | -0.1053 | 0.3215 | |
| GSE29001 | REG1B | 5968 | 205886_at | -0.1591 | 0.1672 | |
| GSE38129 | REG1B | 5968 | 205886_at | -0.0039 | 0.9585 | |
| GSE45670 | REG1B | 5968 | 205886_at | 0.1428 | 0.0660 | |
| GSE53622 | REG1B | 5968 | 42992 | 0.7144 | 0.0000 | |
| GSE53624 | REG1B | 5968 | 42992 | 0.4329 | 0.0085 | |
| GSE63941 | REG1B | 5968 | 205886_at | -0.0610 | 0.6645 | |
| GSE77861 | REG1B | 5968 | 205886_at | -0.1698 | 0.0672 | |
| TCGA | REG1B | 5968 | RNAseq | -1.7951 | 0.4384 |
Upregulated datasets: 0; Downregulated datasets: 0.
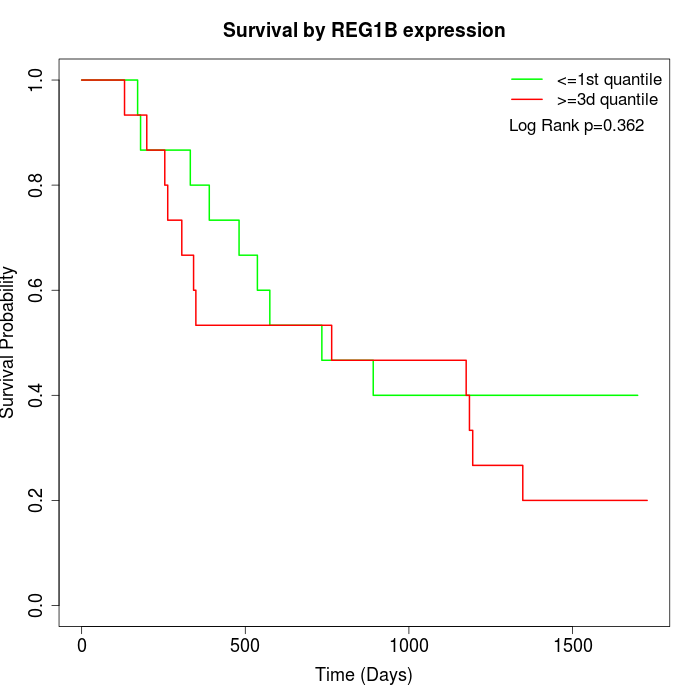
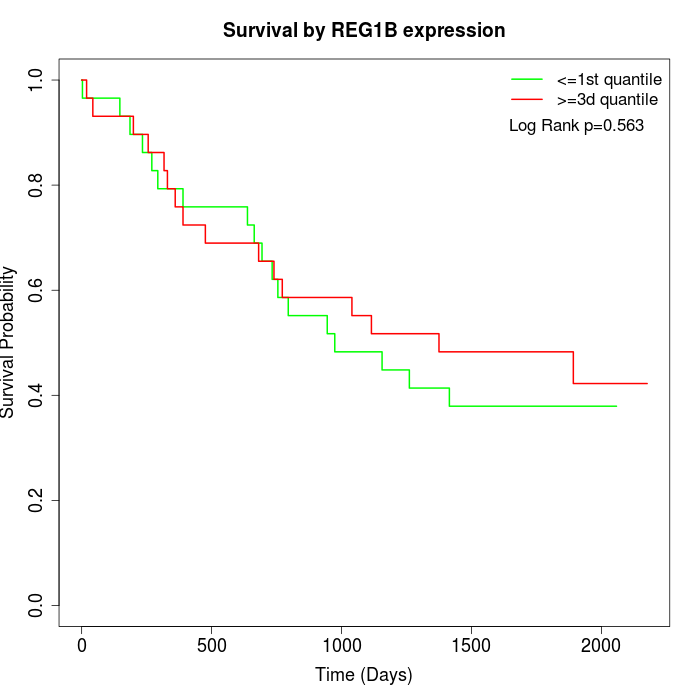
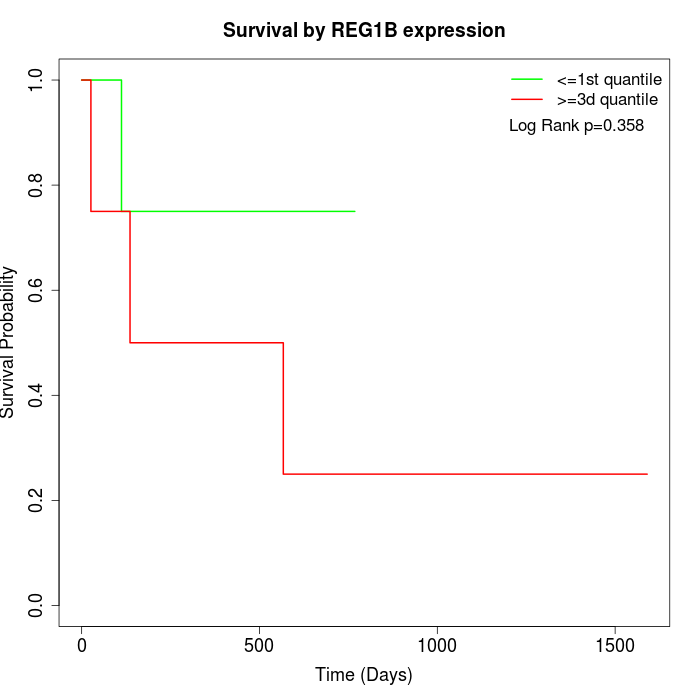
Survival by REG1B expression:
Note: Click image to view full size file.
Copy number change of REG1B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | REG1B | 5968 | 9 | 2 | 19 | |
| GSE20123 | REG1B | 5968 | 8 | 2 | 20 | |
| GSE43470 | REG1B | 5968 | 4 | 0 | 39 | |
| GSE46452 | REG1B | 5968 | 2 | 3 | 54 | |
| GSE47630 | REG1B | 5968 | 7 | 0 | 33 | |
| GSE54993 | REG1B | 5968 | 0 | 6 | 64 | |
| GSE54994 | REG1B | 5968 | 10 | 0 | 43 | |
| GSE60625 | REG1B | 5968 | 0 | 3 | 8 | |
| GSE74703 | REG1B | 5968 | 4 | 0 | 32 | |
| GSE74704 | REG1B | 5968 | 5 | 1 | 14 | |
| TCGA | REG1B | 5968 | 36 | 3 | 57 |
Total number of gains: 85; Total number of losses: 20; Total Number of normals: 383.
Somatic mutations of REG1B:
Generating mutation plots.
Highly correlated genes for REG1B:
Showing top 20/72 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| REG1B | KLK14 | 0.638907 | 3 | 0 | 3 |
| REG1B | SEMA4G | 0.619107 | 3 | 0 | 3 |
| REG1B | AGXT | 0.600682 | 4 | 0 | 4 |
| REG1B | FAM124B | 0.597754 | 4 | 0 | 3 |
| REG1B | LRRC3 | 0.595981 | 3 | 0 | 3 |
| REG1B | MC3R | 0.595697 | 3 | 0 | 3 |
| REG1B | GPHB5 | 0.593105 | 3 | 0 | 3 |
| REG1B | ACVRL1 | 0.582803 | 4 | 0 | 3 |
| REG1B | HSD3B1 | 0.57805 | 4 | 0 | 3 |
| REG1B | PRPS1L1 | 0.577942 | 3 | 0 | 3 |
| REG1B | GJA3 | 0.577478 | 4 | 0 | 3 |
| REG1B | PSPN | 0.571712 | 4 | 0 | 3 |
| REG1B | RPGRIP1 | 0.566148 | 3 | 0 | 3 |
| REG1B | KRTAP11-1 | 0.566001 | 3 | 0 | 3 |
| REG1B | PPP5C | 0.563433 | 4 | 0 | 3 |
| REG1B | PPY | 0.559775 | 4 | 0 | 3 |
| REG1B | DEFA4 | 0.557057 | 4 | 0 | 3 |
| REG1B | MOGAT2 | 0.552039 | 5 | 0 | 3 |
| REG1B | GABRB2 | 0.551774 | 5 | 0 | 3 |
| REG1B | GPR17 | 0.551571 | 4 | 0 | 3 |
For details and further investigation, click here