| Full name: ring finger protein 220 | Alias Symbol: FLJ10597 | ||
| Type: protein-coding gene | Cytoband: 1p34.1 | ||
| Entrez ID: 55182 | HGNC ID: HGNC:25552 | Ensembl Gene: ENSG00000187147 | OMIM ID: 616136 |
| Drug and gene relationship at DGIdb | |||
Expression of RNF220:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RNF220 | 55182 | 219988_s_at | 0.4797 | 0.2497 | |
| GSE20347 | RNF220 | 55182 | 219988_s_at | 0.4221 | 0.0002 | |
| GSE23400 | RNF220 | 55182 | 219988_s_at | 0.3190 | 0.0000 | |
| GSE26886 | RNF220 | 55182 | 219988_s_at | 0.3582 | 0.0534 | |
| GSE29001 | RNF220 | 55182 | 219988_s_at | 0.3672 | 0.2304 | |
| GSE38129 | RNF220 | 55182 | 219988_s_at | 0.3836 | 0.0001 | |
| GSE45670 | RNF220 | 55182 | 219988_s_at | 0.0881 | 0.3813 | |
| GSE53622 | RNF220 | 55182 | 72944 | 0.1579 | 0.0015 | |
| GSE53624 | RNF220 | 55182 | 72944 | 0.1685 | 0.0009 | |
| GSE63941 | RNF220 | 55182 | 219988_s_at | 0.6367 | 0.0553 | |
| GSE77861 | RNF220 | 55182 | 219988_s_at | 0.3722 | 0.1682 | |
| GSE97050 | RNF220 | 55182 | A_33_P3414851 | -0.1119 | 0.6699 | |
| SRP007169 | RNF220 | 55182 | RNAseq | 0.2744 | 0.4697 | |
| SRP008496 | RNF220 | 55182 | RNAseq | 0.2775 | 0.1877 | |
| SRP064894 | RNF220 | 55182 | RNAseq | 0.7102 | 0.0001 | |
| SRP133303 | RNF220 | 55182 | RNAseq | 0.2480 | 0.0849 | |
| SRP159526 | RNF220 | 55182 | RNAseq | 0.3822 | 0.1180 | |
| SRP193095 | RNF220 | 55182 | RNAseq | 0.2994 | 0.0098 | |
| SRP219564 | RNF220 | 55182 | RNAseq | 0.4524 | 0.1823 | |
| TCGA | RNF220 | 55182 | RNAseq | 0.1277 | 0.0094 |
Upregulated datasets: 0; Downregulated datasets: 0.
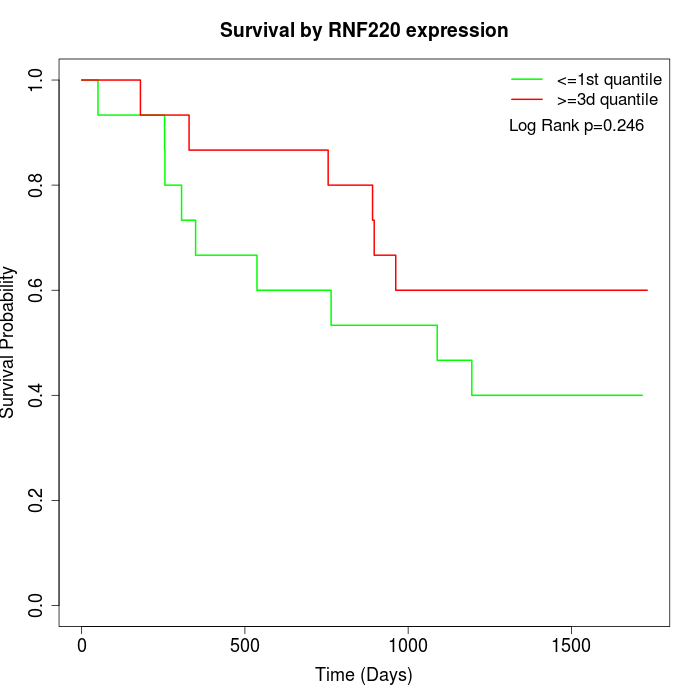
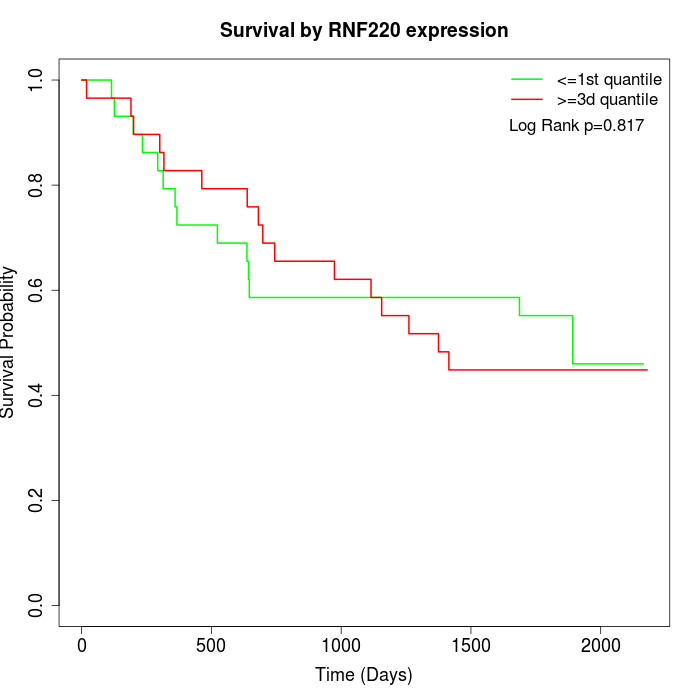
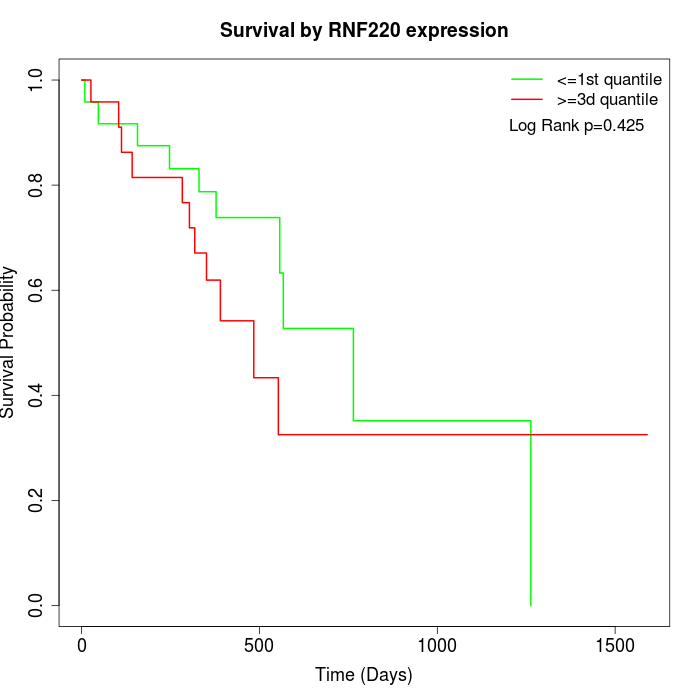
Survival by RNF220 expression:
Note: Click image to view full size file.
Copy number change of RNF220:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RNF220 | 55182 | 4 | 3 | 23 | |
| GSE20123 | RNF220 | 55182 | 4 | 2 | 24 | |
| GSE43470 | RNF220 | 55182 | 7 | 2 | 34 | |
| GSE46452 | RNF220 | 55182 | 2 | 1 | 56 | |
| GSE47630 | RNF220 | 55182 | 9 | 3 | 28 | |
| GSE54993 | RNF220 | 55182 | 0 | 1 | 69 | |
| GSE54994 | RNF220 | 55182 | 13 | 2 | 38 | |
| GSE60625 | RNF220 | 55182 | 0 | 0 | 11 | |
| GSE74703 | RNF220 | 55182 | 6 | 1 | 29 | |
| GSE74704 | RNF220 | 55182 | 2 | 0 | 18 | |
| TCGA | RNF220 | 55182 | 14 | 17 | 65 |
Total number of gains: 61; Total number of losses: 32; Total Number of normals: 395.
Somatic mutations of RNF220:
Generating mutation plots.
Highly correlated genes for RNF220:
Showing top 20/1125 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RNF220 | PACS1 | 0.780479 | 3 | 0 | 3 |
| RNF220 | KLHL36 | 0.765107 | 3 | 0 | 3 |
| RNF220 | NACC1 | 0.760893 | 3 | 0 | 3 |
| RNF220 | EXOC6 | 0.754656 | 3 | 0 | 3 |
| RNF220 | DNAJC19 | 0.752312 | 3 | 0 | 3 |
| RNF220 | THAP5 | 0.739078 | 3 | 0 | 3 |
| RNF220 | ZNF664 | 0.73777 | 3 | 0 | 3 |
| RNF220 | IFNAR1 | 0.735864 | 3 | 0 | 3 |
| RNF220 | ZNF473 | 0.714599 | 3 | 0 | 3 |
| RNF220 | KLHDC3 | 0.713526 | 5 | 0 | 5 |
| RNF220 | ELAVL1 | 0.710876 | 6 | 0 | 5 |
| RNF220 | MAP1S | 0.708365 | 4 | 0 | 4 |
| RNF220 | TMC7 | 0.706191 | 3 | 0 | 3 |
| RNF220 | LNP1 | 0.704127 | 3 | 0 | 3 |
| RNF220 | ASB2 | 0.699163 | 3 | 0 | 3 |
| RNF220 | TUBA1B | 0.697886 | 9 | 0 | 7 |
| RNF220 | DEDD | 0.692343 | 4 | 0 | 3 |
| RNF220 | SYNGR2 | 0.689914 | 5 | 0 | 4 |
| RNF220 | ERI3 | 0.68941 | 12 | 0 | 9 |
| RNF220 | DYNLRB1 | 0.685495 | 4 | 0 | 3 |
For details and further investigation, click here