| Full name: ribonucleotide reductase regulatory subunit M2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 2p25.1 | ||
| Entrez ID: 6241 | HGNC ID: HGNC:10452 | Ensembl Gene: ENSG00000171848 | OMIM ID: 180390 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
RRM2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04115 | p53 signaling pathway |
Expression of RRM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RRM2 | 6241 | 201890_at | 1.0091 | 0.2657 | |
| GSE20347 | RRM2 | 6241 | 201890_at | 1.4000 | 0.0000 | |
| GSE23400 | RRM2 | 6241 | 209773_s_at | 1.3123 | 0.0000 | |
| GSE26886 | RRM2 | 6241 | 201890_at | -0.1922 | 0.7196 | |
| GSE29001 | RRM2 | 6241 | 201890_at | 1.3199 | 0.0008 | |
| GSE38129 | RRM2 | 6241 | 209773_s_at | 1.7254 | 0.0000 | |
| GSE45670 | RRM2 | 6241 | 209773_s_at | 1.4099 | 0.0000 | |
| GSE53622 | RRM2 | 6241 | 86019 | 1.2659 | 0.0000 | |
| GSE53624 | RRM2 | 6241 | 86019 | 1.0737 | 0.0000 | |
| GSE63941 | RRM2 | 6241 | 209773_s_at | 2.5750 | 0.0055 | |
| GSE77861 | RRM2 | 6241 | 209773_s_at | 0.3699 | 0.3047 | |
| GSE97050 | RRM2 | 6241 | A_24_P225616 | 0.4350 | 0.2049 | |
| SRP007169 | RRM2 | 6241 | RNAseq | 1.9993 | 0.0004 | |
| SRP008496 | RRM2 | 6241 | RNAseq | 1.7866 | 0.0000 | |
| SRP064894 | RRM2 | 6241 | RNAseq | 1.2574 | 0.0002 | |
| SRP133303 | RRM2 | 6241 | RNAseq | 1.4345 | 0.0000 | |
| SRP159526 | RRM2 | 6241 | RNAseq | 1.2303 | 0.0002 | |
| SRP193095 | RRM2 | 6241 | RNAseq | 0.8030 | 0.0000 | |
| SRP219564 | RRM2 | 6241 | RNAseq | 1.3684 | 0.0671 | |
| TCGA | RRM2 | 6241 | RNAseq | 1.0332 | 0.0000 |
Upregulated datasets: 14; Downregulated datasets: 0.
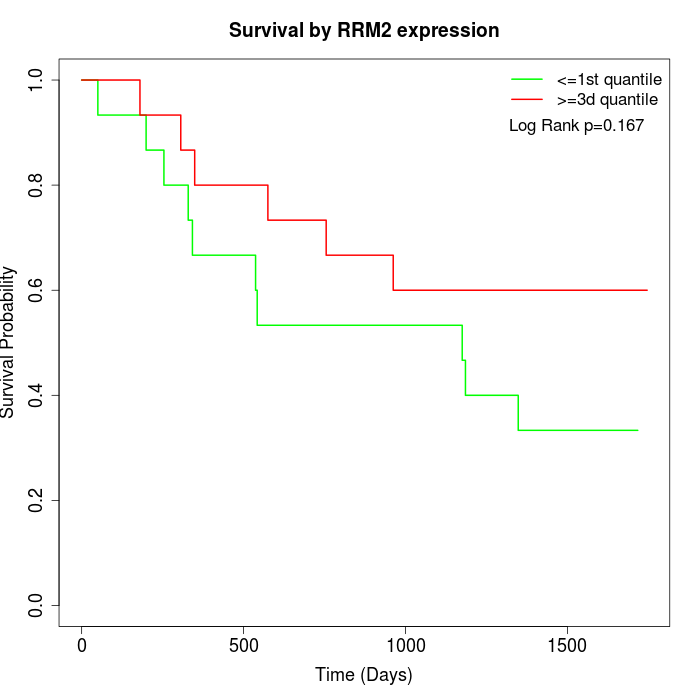
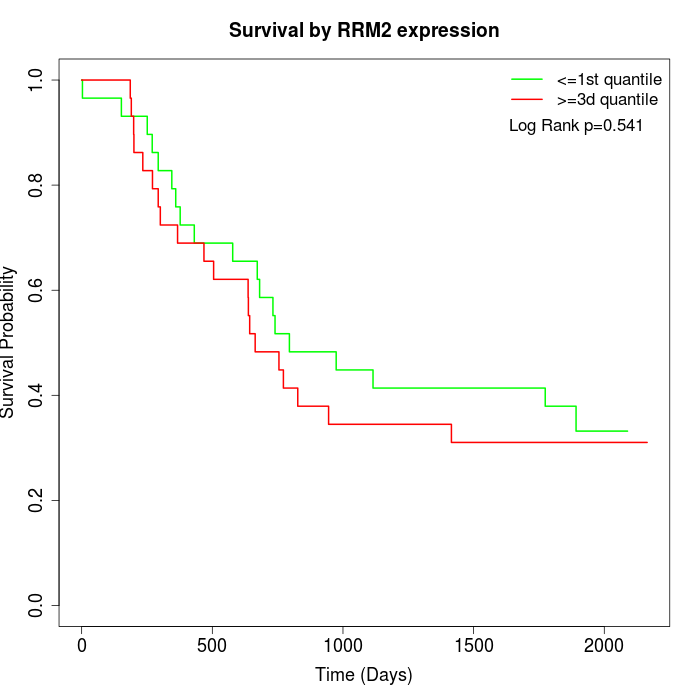
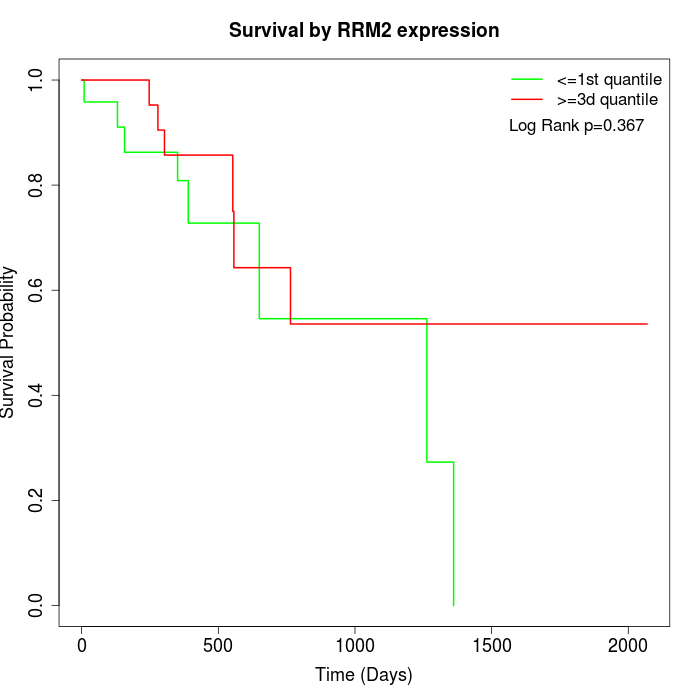
Survival by RRM2 expression:
Note: Click image to view full size file.
Copy number change of RRM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RRM2 | 6241 | 11 | 2 | 17 | |
| GSE20123 | RRM2 | 6241 | 11 | 2 | 17 | |
| GSE43470 | RRM2 | 6241 | 4 | 1 | 38 | |
| GSE46452 | RRM2 | 6241 | 3 | 4 | 52 | |
| GSE47630 | RRM2 | 6241 | 7 | 0 | 33 | |
| GSE54993 | RRM2 | 6241 | 0 | 7 | 63 | |
| GSE54994 | RRM2 | 6241 | 11 | 0 | 42 | |
| GSE60625 | RRM2 | 6241 | 0 | 3 | 8 | |
| GSE74703 | RRM2 | 6241 | 3 | 0 | 33 | |
| GSE74704 | RRM2 | 6241 | 9 | 1 | 10 | |
| TCGA | RRM2 | 6241 | 34 | 5 | 57 |
Total number of gains: 93; Total number of losses: 25; Total Number of normals: 370.
Somatic mutations of RRM2:
Generating mutation plots.
Highly correlated genes for RRM2:
Showing top 20/641 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RRM2 | CDC20 | 0.79482 | 12 | 0 | 12 |
| RRM2 | BUB1B | 0.79092 | 10 | 0 | 9 |
| RRM2 | TTK | 0.780896 | 10 | 0 | 10 |
| RRM2 | MAD2L1 | 0.777502 | 11 | 0 | 11 |
| RRM2 | HMMR | 0.761683 | 11 | 0 | 11 |
| RRM2 | ZWINT | 0.758088 | 12 | 0 | 11 |
| RRM2 | CCNA2 | 0.754735 | 11 | 0 | 10 |
| RRM2 | MCM4 | 0.752587 | 9 | 0 | 9 |
| RRM2 | CDK1 | 0.75064 | 11 | 0 | 10 |
| RRM2 | MELK | 0.750136 | 12 | 0 | 11 |
| RRM2 | CCNB1 | 0.749986 | 11 | 0 | 11 |
| RRM2 | PRC1 | 0.749932 | 11 | 0 | 10 |
| RRM2 | FEN1 | 0.747105 | 10 | 0 | 8 |
| RRM2 | BIRC5 | 0.746402 | 11 | 0 | 10 |
| RRM2 | DLGAP5 | 0.746042 | 13 | 0 | 12 |
| RRM2 | PBK | 0.745118 | 11 | 0 | 10 |
| RRM2 | OIP5 | 0.744546 | 10 | 0 | 9 |
| RRM2 | KIF20A | 0.744119 | 11 | 0 | 10 |
| RRM2 | CDCA8 | 0.741116 | 11 | 0 | 10 |
| RRM2 | NUSAP1 | 0.740272 | 11 | 0 | 9 |
For details and further investigation, click here