| Full name: small EDRK-rich factor 2 | Alias Symbol: H4F5rel|4F5REL|FAM2C|HsT17089 | ||
| Type: protein-coding gene | Cytoband: 15q15.3 | ||
| Entrez ID: 10169 | HGNC ID: HGNC:10757 | Ensembl Gene: ENSG00000140264 | OMIM ID: 605054 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of SERF2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | SERF2 | 10169 | 52444 | -0.3411 | 0.0000 | |
| GSE53624 | SERF2 | 10169 | 52444 | -0.2765 | 0.0000 | |
| GSE97050 | SERF2 | 10169 | A_23_P106299 | -0.3663 | 0.1993 | |
| SRP007169 | SERF2 | 10169 | RNAseq | 0.7348 | 0.0645 | |
| SRP008496 | SERF2 | 10169 | RNAseq | 0.5428 | 0.1232 | |
| SRP064894 | SERF2 | 10169 | RNAseq | 0.2646 | 0.4959 | |
| SRP133303 | SERF2 | 10169 | RNAseq | -0.3114 | 0.0095 | |
| SRP159526 | SERF2 | 10169 | RNAseq | -0.3070 | 0.1672 | |
| SRP193095 | SERF2 | 10169 | RNAseq | -0.5809 | 0.0000 | |
| SRP219564 | SERF2 | 10169 | RNAseq | -0.3808 | 0.2657 | |
| TCGA | SERF2 | 10169 | RNAseq | -0.0789 | 0.0722 |
Upregulated datasets: 0; Downregulated datasets: 0.
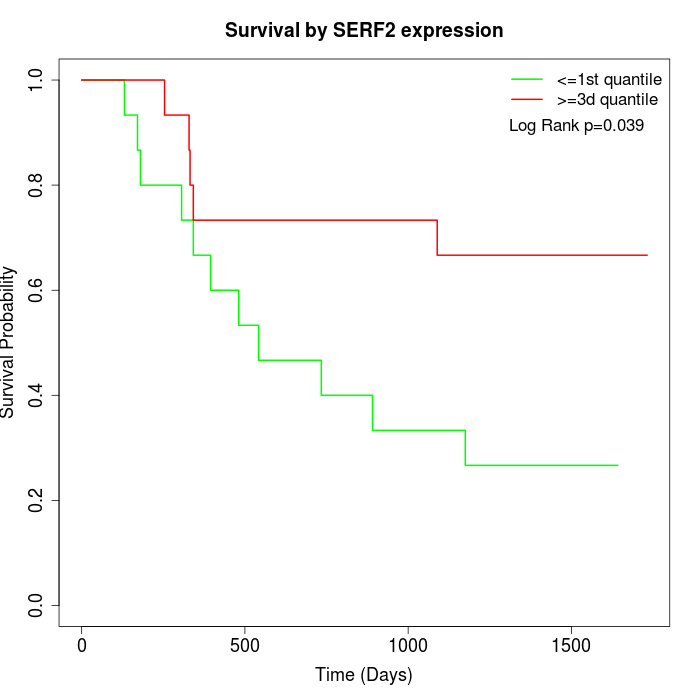
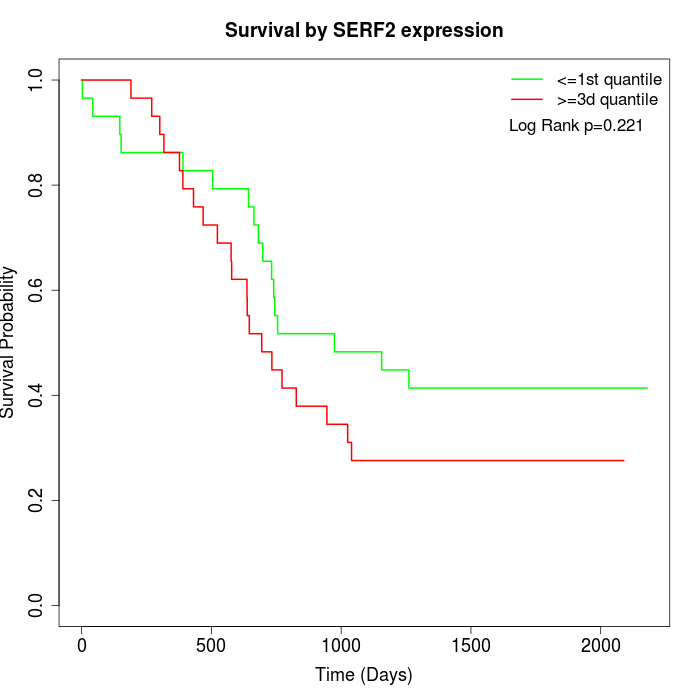
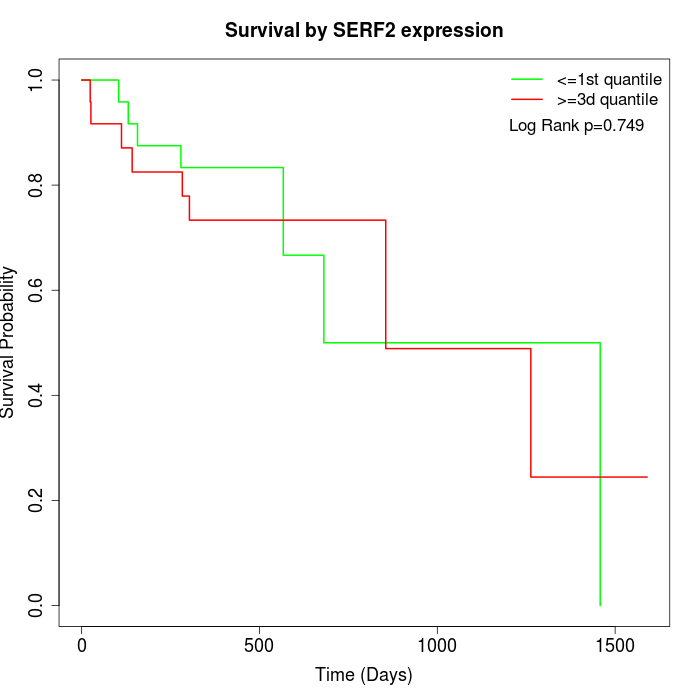
Survival by SERF2 expression:
Note: Click image to view full size file.
Copy number change of SERF2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SERF2 | 10169 | 3 | 4 | 23 | |
| GSE20123 | SERF2 | 10169 | 3 | 5 | 22 | |
| GSE43470 | SERF2 | 10169 | 3 | 4 | 36 | |
| GSE46452 | SERF2 | 10169 | 3 | 7 | 49 | |
| GSE47630 | SERF2 | 10169 | 8 | 10 | 22 | |
| GSE54993 | SERF2 | 10169 | 4 | 6 | 60 | |
| GSE54994 | SERF2 | 10169 | 5 | 7 | 41 | |
| GSE60625 | SERF2 | 10169 | 4 | 0 | 7 | |
| GSE74703 | SERF2 | 10169 | 3 | 3 | 30 | |
| GSE74704 | SERF2 | 10169 | 2 | 2 | 16 | |
| TCGA | SERF2 | 10169 | 12 | 17 | 67 |
Total number of gains: 50; Total number of losses: 65; Total Number of normals: 373.
Somatic mutations of SERF2:
Generating mutation plots.
Highly correlated genes for SERF2:
Showing top 20/26 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SERF2 | SRP14 | 0.734129 | 4 | 0 | 4 |
| SERF2 | COX5B | 0.719522 | 3 | 0 | 3 |
| SERF2 | SPG11 | 0.681331 | 3 | 0 | 3 |
| SERF2 | TMEM109 | 0.679291 | 3 | 0 | 3 |
| SERF2 | COX6B1 | 0.668535 | 4 | 0 | 3 |
| SERF2 | NDUFB2 | 0.665283 | 3 | 0 | 3 |
| SERF2 | WASF2 | 0.664026 | 4 | 0 | 3 |
| SERF2 | RPS9 | 0.657215 | 4 | 0 | 3 |
| SERF2 | HSDL2 | 0.655313 | 3 | 0 | 3 |
| SERF2 | COX6A1 | 0.639218 | 3 | 0 | 3 |
| SERF2 | GUK1 | 0.636492 | 4 | 0 | 3 |
| SERF2 | NDUFAF1 | 0.635769 | 4 | 0 | 3 |
| SERF2 | DPP8 | 0.632876 | 3 | 0 | 3 |
| SERF2 | EDF1 | 0.629803 | 4 | 0 | 3 |
| SERF2 | ALKBH7 | 0.62217 | 4 | 0 | 3 |
| SERF2 | BLOC1S1 | 0.601405 | 4 | 0 | 3 |
| SERF2 | CHMP2A | 0.597403 | 4 | 0 | 3 |
| SERF2 | CAPNS1 | 0.58916 | 4 | 0 | 3 |
| SERF2 | CST3 | 0.5862 | 4 | 0 | 3 |
| SERF2 | COX7A2 | 0.561055 | 4 | 0 | 3 |
For details and further investigation, click here