| Full name: survival of motor neuron 2, centromeric | Alias Symbol: BCD541|SMNC|GEMIN1|TDRD16B | ||
| Type: protein-coding gene | Cytoband: 5q13.2 | ||
| Entrez ID: 6607 | HGNC ID: HGNC:11118 | Ensembl Gene: ENSG00000205571 | OMIM ID: 601627 |
| Drug and gene relationship at DGIdb | |||
Expression of SMN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | SMN2 | 6607 | 106342 | -0.5441 | 0.0028 | |
| GSE53624 | SMN2 | 6607 | 106342 | -0.4733 | 0.0039 | |
| GSE97050 | SMN2 | 6607 | A_24_P126682 | 0.4132 | 0.2086 | |
| SRP007169 | SMN2 | 6607 | RNAseq | 0.7635 | 0.1453 | |
| SRP064894 | SMN2 | 6607 | RNAseq | -0.1644 | 0.6013 | |
| SRP133303 | SMN2 | 6607 | RNAseq | 0.1632 | 0.8406 | |
| SRP159526 | SMN2 | 6607 | RNAseq | 0.1271 | 0.8381 | |
| SRP193095 | SMN2 | 6607 | RNAseq | 0.0618 | 0.7857 | |
| SRP219564 | SMN2 | 6607 | RNAseq | -0.0673 | 0.8426 | |
| TCGA | SMN2 | 6607 | RNAseq | 0.1694 | 0.0031 |
Upregulated datasets: 0; Downregulated datasets: 0.
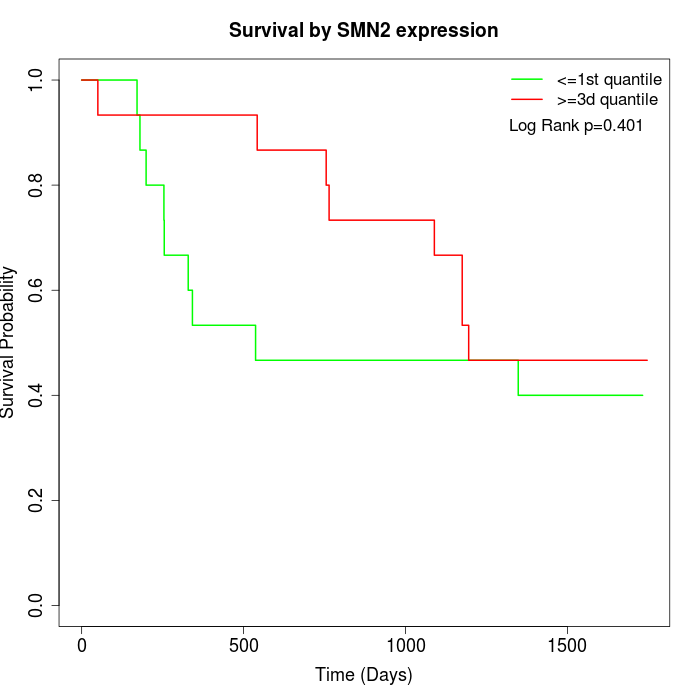
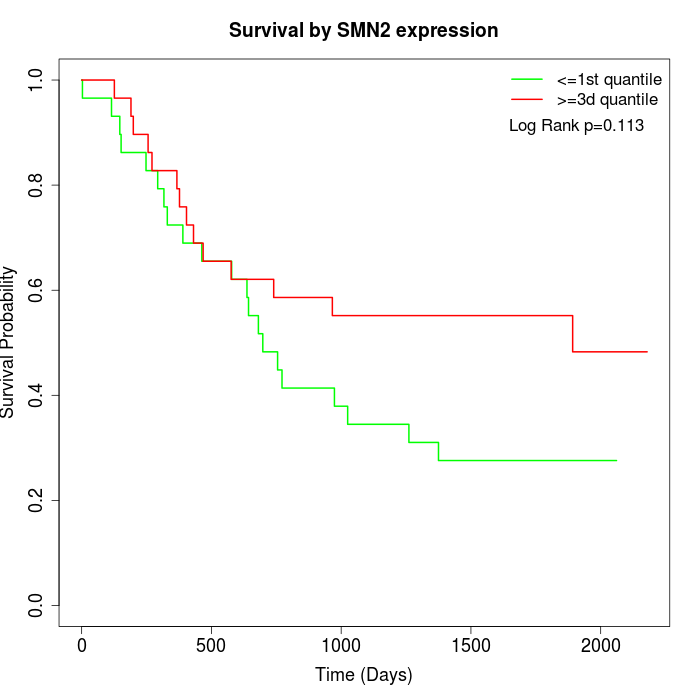
Survival by SMN2 expression:
Note: Click image to view full size file.
Copy number change of SMN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SMN2 | 6607 | 1 | 13 | 16 | |
| GSE20123 | SMN2 | 6607 | 1 | 13 | 16 | |
| GSE43470 | SMN2 | 6607 | 1 | 12 | 30 | |
| GSE46452 | SMN2 | 6607 | 0 | 28 | 31 | |
| GSE47630 | SMN2 | 6607 | 1 | 19 | 20 | |
| GSE54993 | SMN2 | 6607 | 8 | 1 | 61 | |
| GSE54994 | SMN2 | 6607 | 2 | 21 | 30 | |
| GSE60625 | SMN2 | 6607 | 0 | 0 | 11 | |
| GSE74703 | SMN2 | 6607 | 1 | 9 | 26 | |
| GSE74704 | SMN2 | 6607 | 1 | 6 | 13 | |
| TCGA | SMN2 | 6607 | 4 | 51 | 41 |
Total number of gains: 20; Total number of losses: 173; Total Number of normals: 295.
Somatic mutations of SMN2:
Generating mutation plots.
Highly correlated genes for SMN2:
Showing top 20/28 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMN2 | ALKBH3 | 0.879065 | 3 | 0 | 3 |
| SMN2 | MINK1 | 0.729813 | 3 | 0 | 3 |
| SMN2 | PPP4R1 | 0.715089 | 3 | 0 | 3 |
| SMN2 | KCNK6 | 0.709186 | 3 | 0 | 3 |
| SMN2 | BDKRB2 | 0.678985 | 3 | 0 | 3 |
| SMN2 | DNHD1 | 0.678561 | 3 | 0 | 3 |
| SMN2 | NEUROG3 | 0.677 | 3 | 0 | 3 |
| SMN2 | FRMD6 | 0.664509 | 3 | 0 | 3 |
| SMN2 | SPTLC1 | 0.652414 | 3 | 0 | 3 |
| SMN2 | SERPINB8 | 0.645669 | 3 | 0 | 3 |
| SMN2 | UBE2G1 | 0.645342 | 3 | 0 | 3 |
| SMN2 | SH3BGRL3 | 0.631012 | 3 | 0 | 3 |
| SMN2 | SLC22A23 | 0.626055 | 3 | 0 | 3 |
| SMN2 | CLIP1 | 0.625994 | 3 | 0 | 3 |
| SMN2 | GSDMC | 0.621521 | 3 | 0 | 3 |
| SMN2 | CERS3 | 0.618366 | 3 | 0 | 3 |
| SMN2 | ASPSCR1 | 0.608181 | 3 | 0 | 3 |
| SMN2 | TWF1 | 0.602375 | 3 | 0 | 3 |
| SMN2 | FAM83A | 0.600975 | 3 | 0 | 3 |
| SMN2 | ENDOD1 | 0.598265 | 3 | 0 | 3 |
For details and further investigation, click here