| Full name: tripartite motif containing 69 | Alias Symbol: Trif|TRIMLESS | ||
| Type: protein-coding gene | Cytoband: 15q15-q21 | ||
| Entrez ID: 140691 | HGNC ID: HGNC:17857 | Ensembl Gene: ENSG00000185880 | OMIM ID: 616017 |
| Drug and gene relationship at DGIdb | |||
Expression of TRIM69:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM69 | 140691 | 1568592_at | -0.1085 | 0.8168 | |
| GSE26886 | TRIM69 | 140691 | 1568592_at | -0.3938 | 0.3359 | |
| GSE45670 | TRIM69 | 140691 | 1568592_at | 0.9496 | 0.0000 | |
| GSE53622 | TRIM69 | 140691 | 117203 | 0.4628 | 0.0000 | |
| GSE53624 | TRIM69 | 140691 | 117203 | 0.4989 | 0.0000 | |
| GSE63941 | TRIM69 | 140691 | 1568592_at | 0.0920 | 0.8869 | |
| GSE77861 | TRIM69 | 140691 | 1568592_at | 0.0679 | 0.8454 | |
| GSE97050 | TRIM69 | 140691 | A_23_P48826 | 0.0570 | 0.7873 | |
| SRP133303 | TRIM69 | 140691 | RNAseq | 0.4017 | 0.2958 | |
| SRP159526 | TRIM69 | 140691 | RNAseq | -0.0568 | 0.8965 | |
| SRP219564 | TRIM69 | 140691 | RNAseq | 0.4758 | 0.4081 | |
| TCGA | TRIM69 | 140691 | RNAseq | 0.5574 | 0.0239 |
Upregulated datasets: 0; Downregulated datasets: 0.
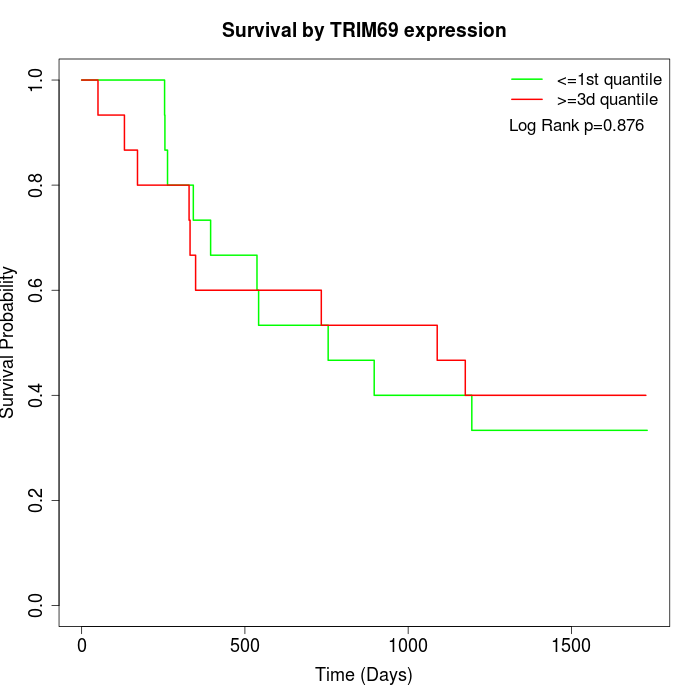
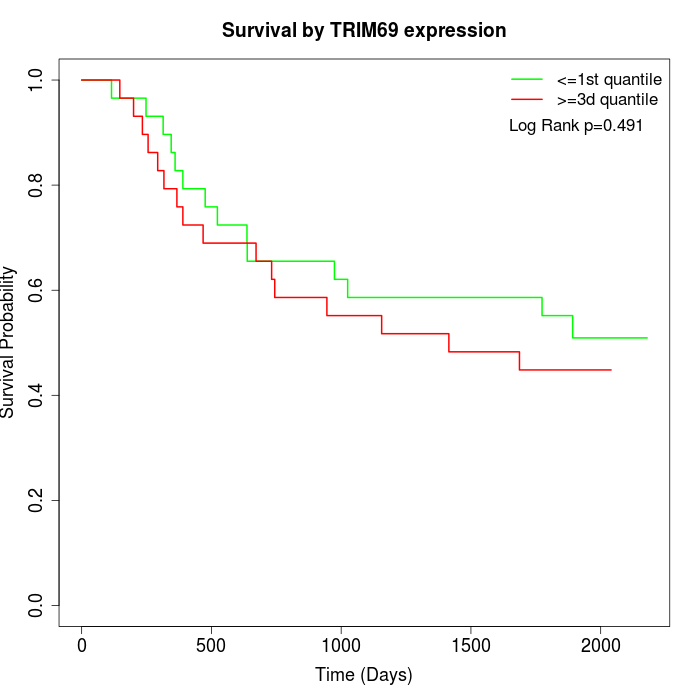
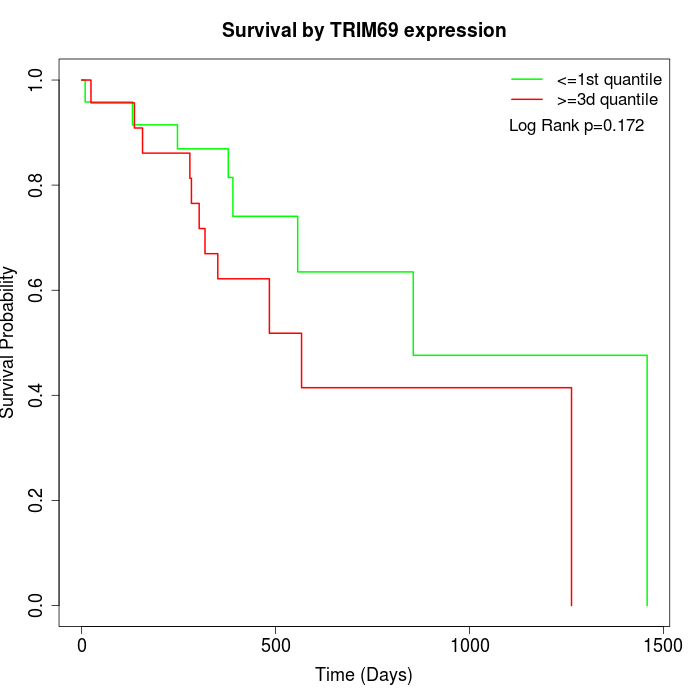
Survival by TRIM69 expression:
Note: Click image to view full size file.
Copy number change of TRIM69:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM69 | 140691 | 3 | 4 | 23 | |
| GSE20123 | TRIM69 | 140691 | 3 | 5 | 22 | |
| GSE43470 | TRIM69 | 140691 | 4 | 4 | 35 | |
| GSE46452 | TRIM69 | 140691 | 3 | 7 | 49 | |
| GSE47630 | TRIM69 | 140691 | 8 | 10 | 22 | |
| GSE54993 | TRIM69 | 140691 | 4 | 6 | 60 | |
| GSE54994 | TRIM69 | 140691 | 5 | 7 | 41 | |
| GSE60625 | TRIM69 | 140691 | 4 | 0 | 7 | |
| GSE74703 | TRIM69 | 140691 | 4 | 3 | 29 | |
| GSE74704 | TRIM69 | 140691 | 1 | 2 | 17 | |
| TCGA | TRIM69 | 140691 | 12 | 16 | 68 |
Total number of gains: 51; Total number of losses: 64; Total Number of normals: 373.
Somatic mutations of TRIM69:
Generating mutation plots.
Highly correlated genes for TRIM69:
Showing top 20/143 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM69 | IFIT3 | 0.697286 | 5 | 0 | 5 |
| TRIM69 | LCP2 | 0.678827 | 3 | 0 | 3 |
| TRIM69 | IFI35 | 0.676862 | 5 | 0 | 5 |
| TRIM69 | CMPK2 | 0.667417 | 6 | 0 | 5 |
| TRIM69 | RSAD2 | 0.667331 | 6 | 0 | 5 |
| TRIM69 | XAF1 | 0.666811 | 5 | 0 | 5 |
| TRIM69 | DHX58 | 0.664397 | 5 | 0 | 5 |
| TRIM69 | LILRB2 | 0.658605 | 3 | 0 | 3 |
| TRIM69 | IFI6 | 0.656002 | 5 | 0 | 4 |
| TRIM69 | AKAP5 | 0.652163 | 3 | 0 | 3 |
| TRIM69 | RNF213 | 0.650471 | 3 | 0 | 3 |
| TRIM69 | FBXO6 | 0.647817 | 5 | 0 | 4 |
| TRIM69 | FCGR3A | 0.643768 | 3 | 0 | 3 |
| TRIM69 | IFIT1 | 0.642953 | 6 | 0 | 5 |
| TRIM69 | SLC7A7 | 0.642104 | 3 | 0 | 3 |
| TRIM69 | PPP1R18 | 0.635729 | 3 | 0 | 3 |
| TRIM69 | TLR1 | 0.633229 | 3 | 0 | 3 |
| TRIM69 | LILRB1 | 0.632682 | 3 | 0 | 3 |
| TRIM69 | SECTM1 | 0.630126 | 4 | 0 | 4 |
| TRIM69 | LILRB4 | 0.626303 | 3 | 0 | 3 |
For details and further investigation, click here