| Full name: ATPase H+ transporting V0 subunit d2 | Alias Symbol: FLJ38708|VMA6 | ||
| Type: protein-coding gene | Cytoband: 8q21.3 | ||
| Entrez ID: 245972 | HGNC ID: HGNC:18266 | Ensembl Gene: ENSG00000147614 | OMIM ID: 618072 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ATP6V0D2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | |
| hsa05152 | Tuberculosis |
Expression of ATP6V0D2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V0D2 | 245972 | 1553153_at | 0.0677 | 0.8473 | |
| GSE26886 | ATP6V0D2 | 245972 | 1553153_at | -0.1216 | 0.2037 | |
| GSE45670 | ATP6V0D2 | 245972 | 1553153_at | 0.1158 | 0.2446 | |
| GSE53622 | ATP6V0D2 | 245972 | 98346 | 2.2879 | 0.0000 | |
| GSE53624 | ATP6V0D2 | 245972 | 98346 | 2.3338 | 0.0000 | |
| GSE63941 | ATP6V0D2 | 245972 | 1553153_at | -0.0975 | 0.7209 | |
| GSE77861 | ATP6V0D2 | 245972 | 1553153_at | 0.0209 | 0.8646 | |
| GSE97050 | ATP6V0D2 | 245972 | A_23_P146146 | 0.2234 | 0.4324 | |
| SRP133303 | ATP6V0D2 | 245972 | RNAseq | 1.1524 | 0.0000 | |
| SRP159526 | ATP6V0D2 | 245972 | RNAseq | 1.2892 | 0.0269 | |
| SRP193095 | ATP6V0D2 | 245972 | RNAseq | 0.6573 | 0.0000 | |
| SRP219564 | ATP6V0D2 | 245972 | RNAseq | 0.8827 | 0.1214 | |
| TCGA | ATP6V0D2 | 245972 | RNAseq | 1.6859 | 0.0000 |
Upregulated datasets: 5; Downregulated datasets: 0.
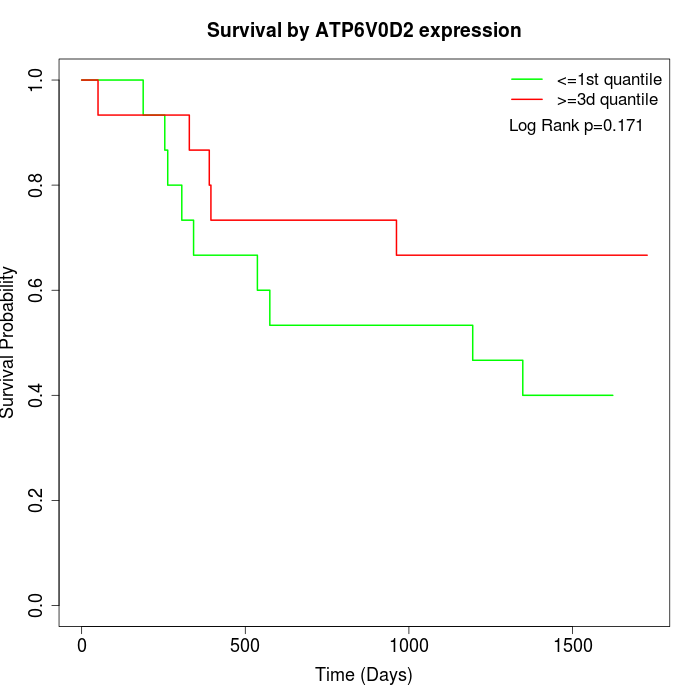
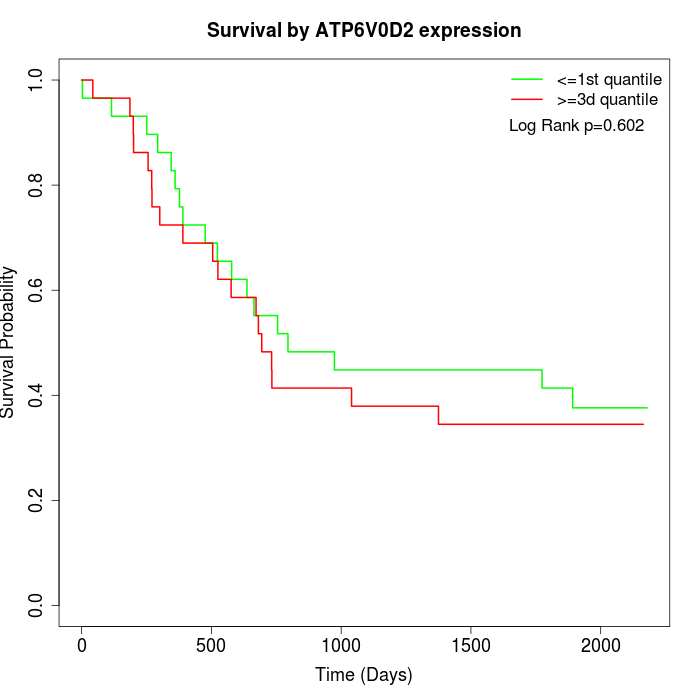
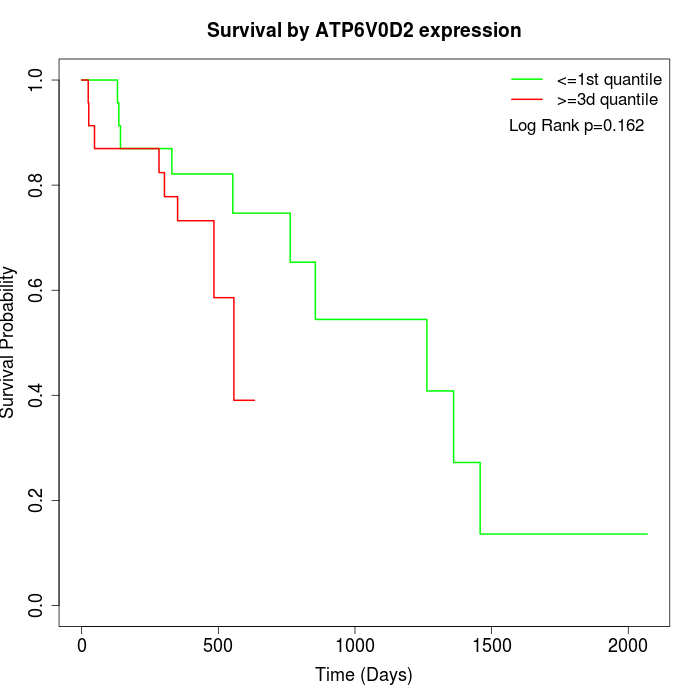
Survival by ATP6V0D2 expression:
Note: Click image to view full size file.
Copy number change of ATP6V0D2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V0D2 | 245972 | 14 | 1 | 15 | |
| GSE20123 | ATP6V0D2 | 245972 | 16 | 1 | 13 | |
| GSE43470 | ATP6V0D2 | 245972 | 19 | 1 | 23 | |
| GSE46452 | ATP6V0D2 | 245972 | 21 | 0 | 38 | |
| GSE47630 | ATP6V0D2 | 245972 | 23 | 0 | 17 | |
| GSE54993 | ATP6V0D2 | 245972 | 0 | 19 | 51 | |
| GSE54994 | ATP6V0D2 | 245972 | 32 | 1 | 20 | |
| GSE60625 | ATP6V0D2 | 245972 | 0 | 4 | 7 | |
| GSE74703 | ATP6V0D2 | 245972 | 16 | 1 | 19 | |
| GSE74704 | ATP6V0D2 | 245972 | 10 | 0 | 10 | |
| TCGA | ATP6V0D2 | 245972 | 54 | 2 | 40 |
Total number of gains: 205; Total number of losses: 30; Total Number of normals: 253.
Somatic mutations of ATP6V0D2:
Generating mutation plots.
Highly correlated genes for ATP6V0D2:
Showing top 20/207 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V0D2 | CHSY3 | 0.770454 | 3 | 0 | 3 |
| ATP6V0D2 | OLR1 | 0.758251 | 3 | 0 | 3 |
| ATP6V0D2 | CTHRC1 | 0.710284 | 3 | 0 | 3 |
| ATP6V0D2 | ANGPT2 | 0.703931 | 3 | 0 | 3 |
| ATP6V0D2 | LAIR2 | 0.695867 | 3 | 0 | 3 |
| ATP6V0D2 | RCN3 | 0.687856 | 3 | 0 | 3 |
| ATP6V0D2 | ZNF469 | 0.683571 | 4 | 0 | 4 |
| ATP6V0D2 | OLFML2B | 0.673012 | 3 | 0 | 3 |
| ATP6V0D2 | CDH11 | 0.670991 | 4 | 0 | 4 |
| ATP6V0D2 | FST | 0.668681 | 4 | 0 | 4 |
| ATP6V0D2 | RGS3 | 0.657932 | 4 | 0 | 4 |
| ATP6V0D2 | FTL | 0.657596 | 3 | 0 | 3 |
| ATP6V0D2 | COL1A1 | 0.651785 | 3 | 0 | 3 |
| ATP6V0D2 | APH1B | 0.651672 | 3 | 0 | 3 |
| ATP6V0D2 | ADSL | 0.649462 | 3 | 0 | 3 |
| ATP6V0D2 | DPP4 | 0.648963 | 4 | 0 | 4 |
| ATP6V0D2 | WDR54 | 0.646736 | 3 | 0 | 3 |
| ATP6V0D2 | KCND2 | 0.646223 | 4 | 0 | 4 |
| ATP6V0D2 | GMPPA | 0.645294 | 3 | 0 | 3 |
| ATP6V0D2 | HMGXB3 | 0.644667 | 3 | 0 | 3 |
For details and further investigation, click here