| Full name: complement C9 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 5p13.1 | ||
| Entrez ID: 735 | HGNC ID: HGNC:1358 | Ensembl Gene: ENSG00000113600 | OMIM ID: 120940 |
| Drug and gene relationship at DGIdb | |||
Expression of C9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | C9 | 735 | 206727_at | 0.0883 | 0.6391 | |
| GSE20347 | C9 | 735 | 206727_at | 0.1186 | 0.0218 | |
| GSE23400 | C9 | 735 | 206727_at | -0.0252 | 0.1941 | |
| GSE26886 | C9 | 735 | 206727_at | 0.2167 | 0.1536 | |
| GSE29001 | C9 | 735 | 206727_at | 0.3783 | 0.1106 | |
| GSE38129 | C9 | 735 | 206727_at | 0.0525 | 0.3234 | |
| GSE45670 | C9 | 735 | 206727_at | 0.1115 | 0.1926 | |
| GSE53622 | C9 | 735 | 7425 | 0.3085 | 0.2191 | |
| GSE53624 | C9 | 735 | 7425 | -0.1563 | 0.5131 | |
| GSE63941 | C9 | 735 | 206727_at | -0.2999 | 0.2427 | |
| GSE77861 | C9 | 735 | 206727_at | -0.0053 | 0.9629 | |
| SRP133303 | C9 | 735 | RNAseq | 0.0074 | 0.9903 | |
| TCGA | C9 | 735 | RNAseq | 2.7214 | 0.0011 |
Upregulated datasets: 1; Downregulated datasets: 0.
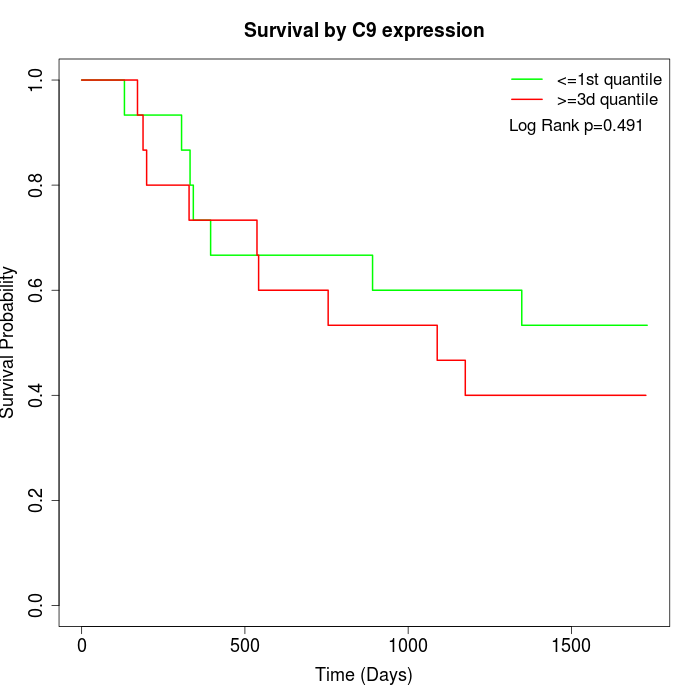
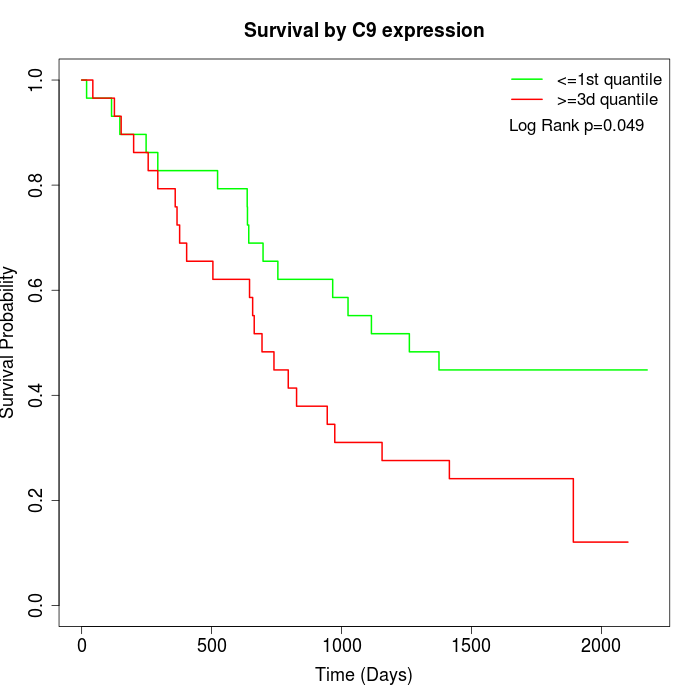
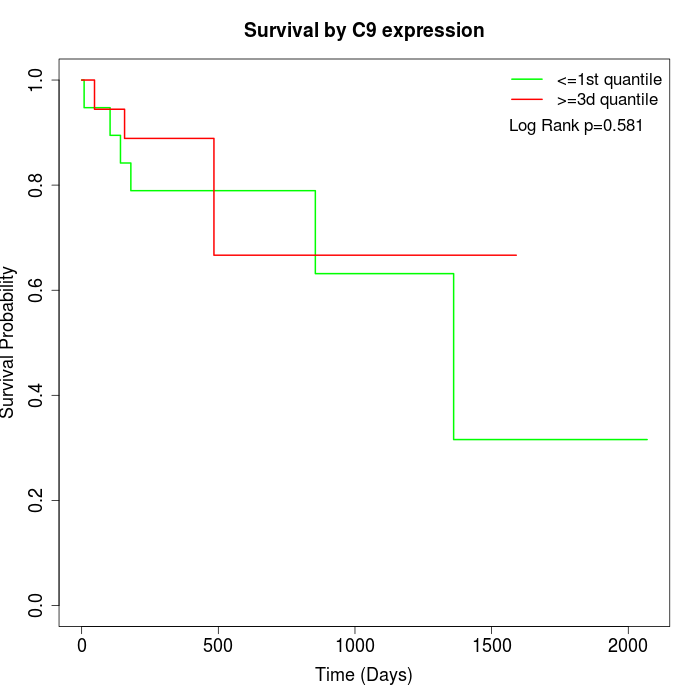
Survival by C9 expression:
Note: Click image to view full size file.
Copy number change of C9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | C9 | 735 | 10 | 0 | 20 | |
| GSE20123 | C9 | 735 | 10 | 0 | 20 | |
| GSE43470 | C9 | 735 | 17 | 0 | 26 | |
| GSE46452 | C9 | 735 | 5 | 22 | 32 | |
| GSE47630 | C9 | 735 | 6 | 12 | 22 | |
| GSE54993 | C9 | 735 | 5 | 4 | 61 | |
| GSE54994 | C9 | 735 | 24 | 2 | 27 | |
| GSE60625 | C9 | 735 | 0 | 0 | 11 | |
| GSE74703 | C9 | 735 | 13 | 0 | 23 | |
| GSE74704 | C9 | 735 | 8 | 0 | 12 | |
| TCGA | C9 | 735 | 52 | 6 | 38 |
Total number of gains: 150; Total number of losses: 46; Total Number of normals: 292.
Somatic mutations of C9:
Generating mutation plots.
Highly correlated genes for C9:
Showing top 20/31 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| C9 | MBL2 | 0.619291 | 4 | 0 | 3 |
| C9 | APOF | 0.608036 | 4 | 0 | 3 |
| C9 | PF4V1 | 0.602483 | 3 | 0 | 3 |
| C9 | DNTT | 0.579087 | 3 | 0 | 3 |
| C9 | DPF3 | 0.577417 | 3 | 0 | 3 |
| C9 | HBQ1 | 0.576266 | 3 | 0 | 3 |
| C9 | FGF20 | 0.574844 | 3 | 0 | 3 |
| C9 | OSMR | 0.573736 | 3 | 0 | 3 |
| C9 | LINC00597 | 0.566488 | 3 | 0 | 3 |
| C9 | MSX1 | 0.552369 | 4 | 0 | 3 |
| C9 | BRF2 | 0.550076 | 4 | 0 | 3 |
| C9 | STIL | 0.548019 | 4 | 0 | 3 |
| C9 | LIN28A | 0.54647 | 6 | 0 | 4 |
| C9 | ADAMTS12 | 0.544064 | 4 | 0 | 3 |
| C9 | GRK4 | 0.543689 | 4 | 0 | 3 |
| C9 | MRGPRX1 | 0.543368 | 4 | 0 | 3 |
| C9 | GRM1 | 0.538103 | 4 | 0 | 3 |
| C9 | LILRA3 | 0.536193 | 5 | 0 | 3 |
| C9 | TRIP13 | 0.534057 | 4 | 0 | 3 |
| C9 | MYF5 | 0.533748 | 3 | 0 | 3 |
For details and further investigation, click here