| Full name: calcium voltage-gated channel auxiliary subunit beta 3 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12q13.12 | ||
| Entrez ID: 784 | HGNC ID: HGNC:1403 | Ensembl Gene: ENSG00000167535 | OMIM ID: 601958 |
| Drug and gene relationship at DGIdb | |||
CACNB3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04261 | Adrenergic signaling in cardiomyocytes | |
| hsa04921 | Oxytocin signaling pathway |
Expression of CACNB3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CACNB3 | 784 | 209530_at | -0.0173 | 0.9864 | |
| GSE20347 | CACNB3 | 784 | 209530_at | -0.3514 | 0.0036 | |
| GSE23400 | CACNB3 | 784 | 209530_at | -0.1226 | 0.0047 | |
| GSE26886 | CACNB3 | 784 | 34726_at | 0.0228 | 0.9093 | |
| GSE29001 | CACNB3 | 784 | 34726_at | -0.1911 | 0.4610 | |
| GSE38129 | CACNB3 | 784 | 209530_at | -0.1938 | 0.1173 | |
| GSE45670 | CACNB3 | 784 | 34726_at | -0.2299 | 0.0559 | |
| GSE53622 | CACNB3 | 784 | 100073 | 0.0873 | 0.2287 | |
| GSE53624 | CACNB3 | 784 | 100073 | 0.0595 | 0.2494 | |
| GSE63941 | CACNB3 | 784 | 209530_at | -0.4890 | 0.2288 | |
| GSE77861 | CACNB3 | 784 | 34726_at | -0.2244 | 0.1907 | |
| GSE97050 | CACNB3 | 784 | A_23_P204016 | -0.0961 | 0.8225 | |
| SRP007169 | CACNB3 | 784 | RNAseq | -1.4841 | 0.0009 | |
| SRP008496 | CACNB3 | 784 | RNAseq | -1.1588 | 0.0003 | |
| SRP064894 | CACNB3 | 784 | RNAseq | 0.1234 | 0.4581 | |
| SRP133303 | CACNB3 | 784 | RNAseq | -0.2988 | 0.1415 | |
| SRP159526 | CACNB3 | 784 | RNAseq | -0.3340 | 0.4343 | |
| SRP193095 | CACNB3 | 784 | RNAseq | -0.4452 | 0.0257 | |
| SRP219564 | CACNB3 | 784 | RNAseq | -0.3220 | 0.1821 | |
| TCGA | CACNB3 | 784 | RNAseq | 0.2220 | 0.0071 |
Upregulated datasets: 0; Downregulated datasets: 2.
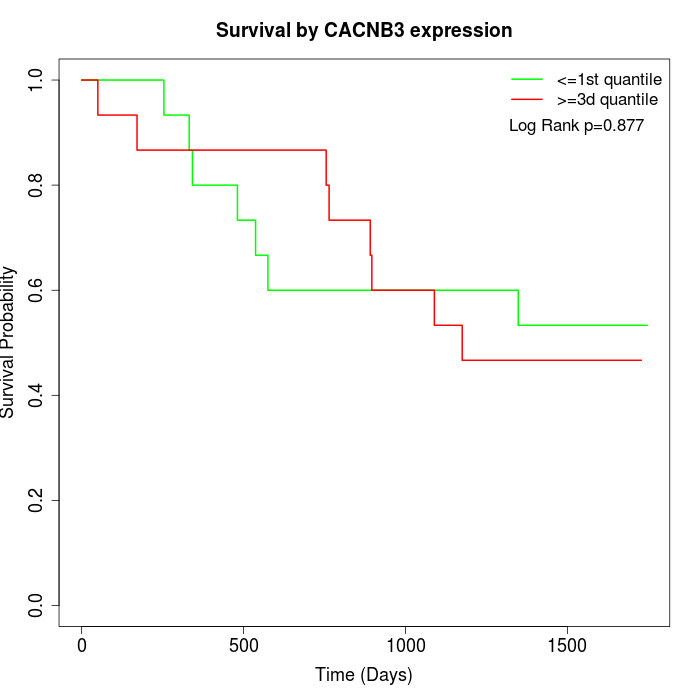
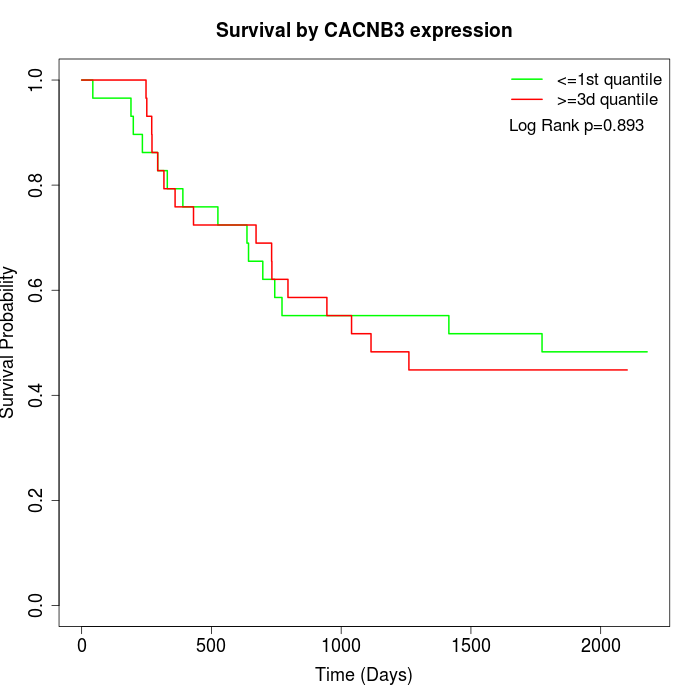
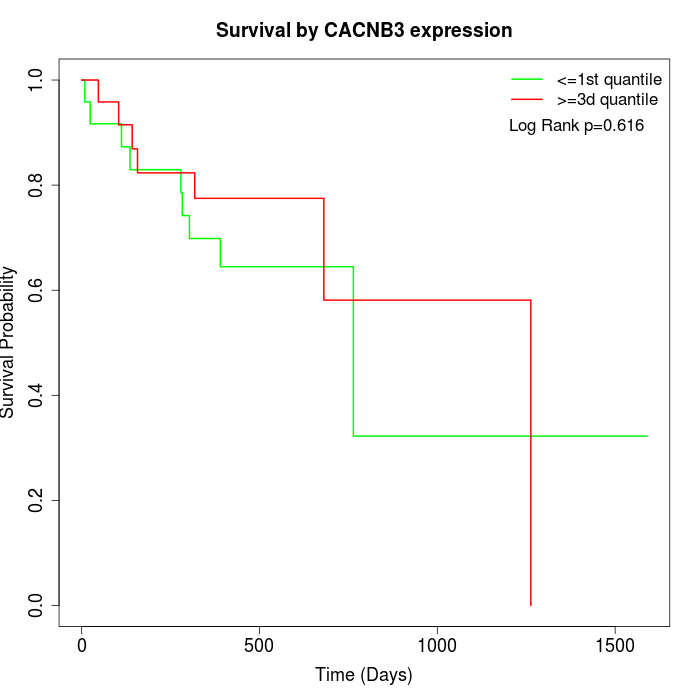
Survival by CACNB3 expression:
Note: Click image to view full size file.
Copy number change of CACNB3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CACNB3 | 784 | 5 | 1 | 24 | |
| GSE20123 | CACNB3 | 784 | 5 | 1 | 24 | |
| GSE43470 | CACNB3 | 784 | 4 | 1 | 38 | |
| GSE46452 | CACNB3 | 784 | 8 | 1 | 50 | |
| GSE47630 | CACNB3 | 784 | 11 | 2 | 27 | |
| GSE54993 | CACNB3 | 784 | 0 | 5 | 65 | |
| GSE54994 | CACNB3 | 784 | 6 | 1 | 46 | |
| GSE60625 | CACNB3 | 784 | 0 | 0 | 11 | |
| GSE74703 | CACNB3 | 784 | 4 | 1 | 31 | |
| GSE74704 | CACNB3 | 784 | 3 | 1 | 16 | |
| TCGA | CACNB3 | 784 | 16 | 12 | 68 |
Total number of gains: 62; Total number of losses: 26; Total Number of normals: 400.
Somatic mutations of CACNB3:
Generating mutation plots.
Highly correlated genes for CACNB3:
Showing top 20/106 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CACNB3 | DXO | 0.740759 | 3 | 0 | 3 |
| CACNB3 | SYCE1L | 0.703797 | 4 | 0 | 4 |
| CACNB3 | HPS1 | 0.699454 | 3 | 0 | 3 |
| CACNB3 | LYPLA2 | 0.668013 | 4 | 0 | 4 |
| CACNB3 | AKR7A3 | 0.662355 | 4 | 0 | 4 |
| CACNB3 | ATF6B | 0.655381 | 4 | 0 | 4 |
| CACNB3 | ABCA2 | 0.654593 | 4 | 0 | 3 |
| CACNB3 | PARS2 | 0.650829 | 3 | 0 | 3 |
| CACNB3 | SMR3A | 0.648469 | 4 | 0 | 3 |
| CACNB3 | PDXK | 0.639175 | 3 | 0 | 3 |
| CACNB3 | PNPLA2 | 0.634691 | 3 | 0 | 3 |
| CACNB3 | ZNF446 | 0.633723 | 3 | 0 | 3 |
| CACNB3 | VIPR2 | 0.631154 | 3 | 0 | 3 |
| CACNB3 | SSH3 | 0.623749 | 3 | 0 | 3 |
| CACNB3 | ZBTB45 | 0.623596 | 4 | 0 | 3 |
| CACNB3 | FAM181A | 0.621329 | 3 | 0 | 3 |
| CACNB3 | AP4M1 | 0.619676 | 3 | 0 | 3 |
| CACNB3 | LIMD1 | 0.617677 | 4 | 0 | 3 |
| CACNB3 | CAPZB | 0.617234 | 5 | 0 | 5 |
| CACNB3 | DSCAM | 0.607426 | 4 | 0 | 3 |
For details and further investigation, click here