| Full name: claudin 17 | Alias Symbol: MGC126552|MGC126554 | ||
| Type: protein-coding gene | Cytoband: 21q21.3 | ||
| Entrez ID: 26285 | HGNC ID: HGNC:2038 | Ensembl Gene: ENSG00000156282 | OMIM ID: 617005 |
| Drug and gene relationship at DGIdb | |||
CLDN17 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04530 | Tight junction |
Expression of CLDN17:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLDN17 | 26285 | 221328_at | -1.0046 | 0.0694 | |
| GSE20347 | CLDN17 | 26285 | 221328_at | -0.8502 | 0.0002 | |
| GSE23400 | CLDN17 | 26285 | 221328_at | -0.6709 | 0.0000 | |
| GSE26886 | CLDN17 | 26285 | 221328_at | -1.1878 | 0.0005 | |
| GSE29001 | CLDN17 | 26285 | 221328_at | -1.1051 | 0.0004 | |
| GSE38129 | CLDN17 | 26285 | 221328_at | -0.8657 | 0.0001 | |
| GSE45670 | CLDN17 | 26285 | 221328_at | -0.6084 | 0.1379 | |
| GSE53622 | CLDN17 | 26285 | 61103 | -3.1603 | 0.0000 | |
| GSE53624 | CLDN17 | 26285 | 61103 | -4.1878 | 0.0000 | |
| GSE63941 | CLDN17 | 26285 | 221328_at | 0.1891 | 0.2915 | |
| GSE77861 | CLDN17 | 26285 | 221328_at | -0.9931 | 0.0270 | |
| SRP007169 | CLDN17 | 26285 | RNAseq | -7.5438 | 0.0000 | |
| SRP008496 | CLDN17 | 26285 | RNAseq | -6.6902 | 0.0000 | |
| SRP064894 | CLDN17 | 26285 | RNAseq | -4.9114 | 0.0000 | |
| SRP133303 | CLDN17 | 26285 | RNAseq | -4.1852 | 0.0000 | |
| SRP159526 | CLDN17 | 26285 | RNAseq | -3.5739 | 0.0010 | |
| TCGA | CLDN17 | 26285 | RNAseq | -0.3976 | 0.6787 |
Upregulated datasets: 0; Downregulated datasets: 9.
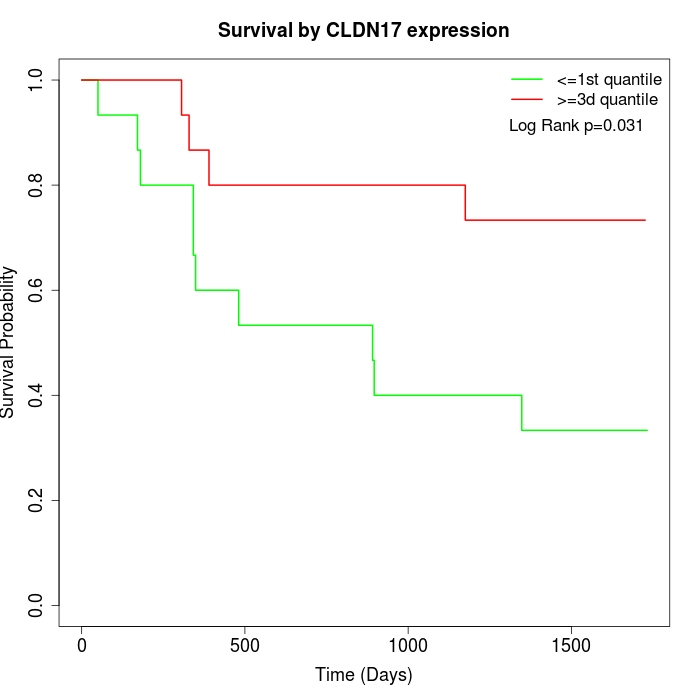
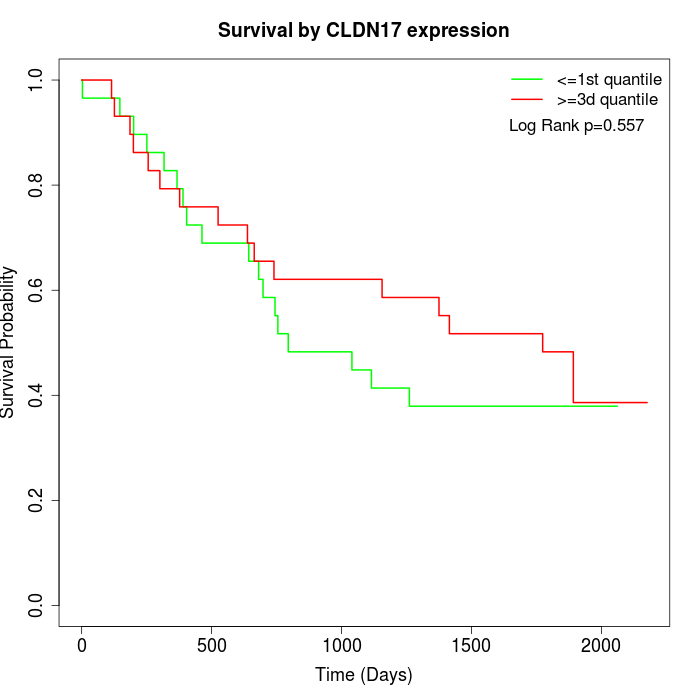
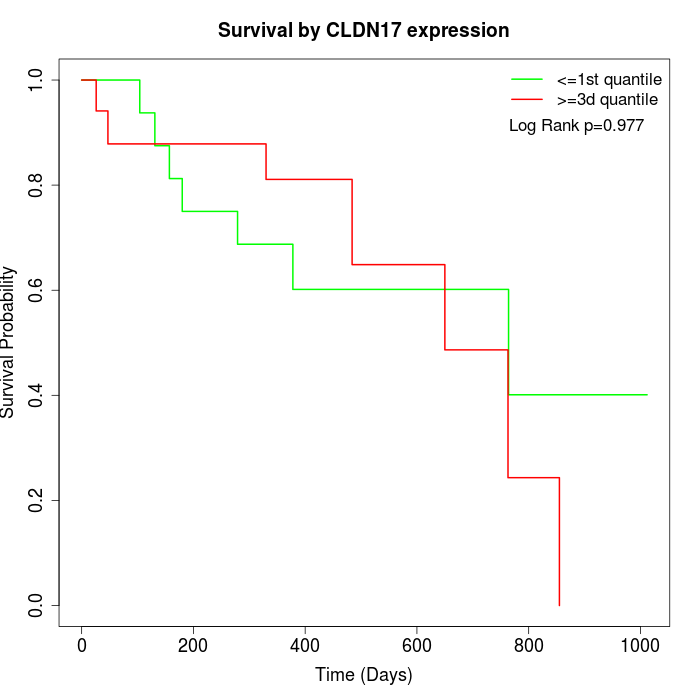
Survival by CLDN17 expression:
Note: Click image to view full size file.
Copy number change of CLDN17:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLDN17 | 26285 | 2 | 10 | 18 | |
| GSE20123 | CLDN17 | 26285 | 2 | 10 | 18 | |
| GSE43470 | CLDN17 | 26285 | 2 | 9 | 32 | |
| GSE46452 | CLDN17 | 26285 | 1 | 21 | 37 | |
| GSE47630 | CLDN17 | 26285 | 6 | 17 | 17 | |
| GSE54993 | CLDN17 | 26285 | 9 | 1 | 60 | |
| GSE54994 | CLDN17 | 26285 | 1 | 9 | 43 | |
| GSE60625 | CLDN17 | 26285 | 0 | 0 | 11 | |
| GSE74703 | CLDN17 | 26285 | 2 | 6 | 28 | |
| GSE74704 | CLDN17 | 26285 | 2 | 6 | 12 | |
| TCGA | CLDN17 | 26285 | 8 | 36 | 52 |
Total number of gains: 35; Total number of losses: 125; Total Number of normals: 328.
Somatic mutations of CLDN17:
Generating mutation plots.
Highly correlated genes for CLDN17:
Showing top 20/1299 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLDN17 | IL36A | 0.810698 | 10 | 0 | 10 |
| CLDN17 | RAET1E | 0.807726 | 6 | 0 | 6 |
| CLDN17 | PRSS2 | 0.796337 | 8 | 0 | 8 |
| CLDN17 | DKK4 | 0.79253 | 10 | 0 | 10 |
| CLDN17 | GDPD3 | 0.78792 | 10 | 0 | 10 |
| CLDN17 | GYS2 | 0.783478 | 10 | 0 | 10 |
| CLDN17 | ENDOU | 0.781771 | 10 | 0 | 10 |
| CLDN17 | HCG22 | 0.773474 | 6 | 0 | 6 |
| CLDN17 | KRT78 | 0.770459 | 6 | 0 | 6 |
| CLDN17 | PRSS27 | 0.769664 | 6 | 0 | 6 |
| CLDN17 | TRIP10 | 0.768181 | 10 | 0 | 10 |
| CLDN17 | BNIPL | 0.766126 | 6 | 0 | 6 |
| CLDN17 | ATG9B | 0.760981 | 6 | 0 | 6 |
| CLDN17 | MAPK3 | 0.759099 | 10 | 0 | 10 |
| CLDN17 | SPINK7 | 0.757838 | 7 | 0 | 6 |
| CLDN17 | CNFN | 0.756454 | 7 | 0 | 6 |
| CLDN17 | SLURP1 | 0.755457 | 11 | 0 | 10 |
| CLDN17 | CRISP3 | 0.7544 | 10 | 0 | 10 |
| CLDN17 | TMPRSS11E | 0.752856 | 10 | 0 | 10 |
| CLDN17 | CRCT1 | 0.752039 | 11 | 0 | 10 |
For details and further investigation, click here