| Full name: eva-1 homolog A, regulator of programmed cell death | Alias Symbol: FLJ13391 | ||
| Type: protein-coding gene | Cytoband: 2p12 | ||
| Entrez ID: 84141 | HGNC ID: HGNC:25816 | Ensembl Gene: ENSG00000115363 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of EVA1A:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EVA1A | 84141 | 227828_s_at | 0.8968 | 0.2880 | |
| GSE26886 | EVA1A | 84141 | 227828_s_at | 0.9882 | 0.0008 | |
| GSE45670 | EVA1A | 84141 | 227828_s_at | 1.0331 | 0.0273 | |
| GSE53622 | EVA1A | 84141 | 57627 | 1.1718 | 0.0000 | |
| GSE53624 | EVA1A | 84141 | 57627 | 1.5113 | 0.0000 | |
| GSE63941 | EVA1A | 84141 | 227828_s_at | -1.9814 | 0.1293 | |
| GSE77861 | EVA1A | 84141 | 227828_s_at | 0.1081 | 0.5179 | |
| SRP007169 | EVA1A | 84141 | RNAseq | 4.1497 | 0.0035 | |
| SRP064894 | EVA1A | 84141 | RNAseq | 3.3887 | 0.0000 | |
| SRP133303 | EVA1A | 84141 | RNAseq | 3.1748 | 0.0000 | |
| SRP219564 | EVA1A | 84141 | RNAseq | 3.2915 | 0.0000 |
Upregulated datasets: 7; Downregulated datasets: 0.
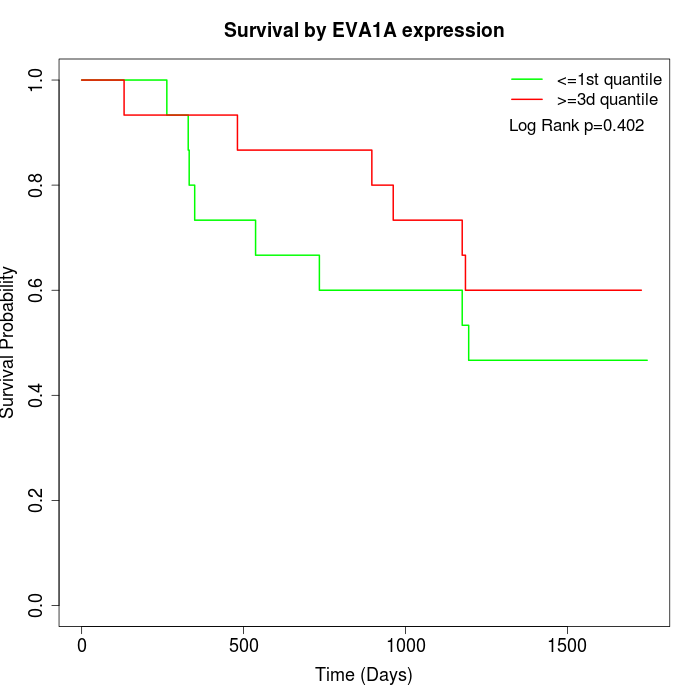
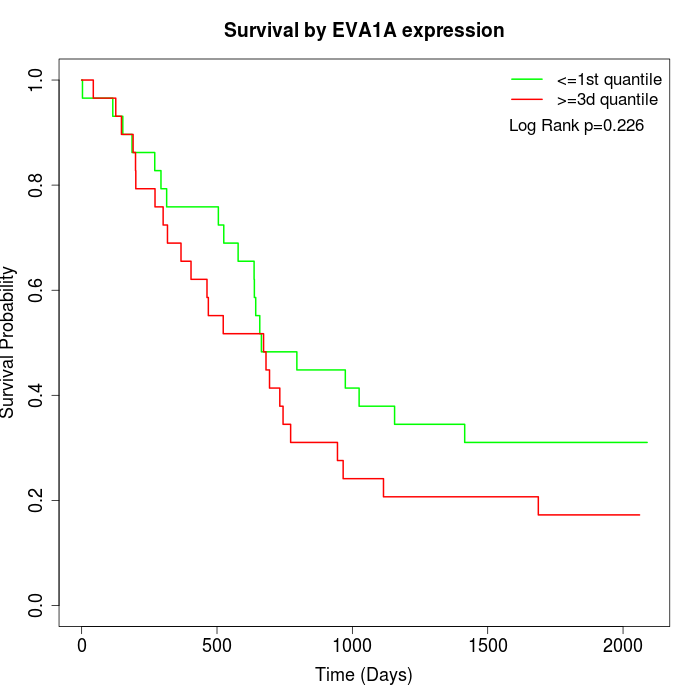
Survival by EVA1A expression:
Note: Click image to view full size file.
Copy number change of EVA1A:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EVA1A | 84141 | 9 | 3 | 18 | |
| GSE20123 | EVA1A | 84141 | 9 | 3 | 18 | |
| GSE43470 | EVA1A | 84141 | 4 | 0 | 39 | |
| GSE46452 | EVA1A | 84141 | 2 | 3 | 54 | |
| GSE47630 | EVA1A | 84141 | 7 | 0 | 33 | |
| GSE54993 | EVA1A | 84141 | 0 | 6 | 64 | |
| GSE54994 | EVA1A | 84141 | 10 | 0 | 43 | |
| GSE60625 | EVA1A | 84141 | 0 | 3 | 8 | |
| GSE74703 | EVA1A | 84141 | 4 | 0 | 32 | |
| GSE74704 | EVA1A | 84141 | 7 | 1 | 12 | |
| TCGA | EVA1A | 84141 | 36 | 2 | 58 |
Total number of gains: 88; Total number of losses: 21; Total Number of normals: 379.
Somatic mutations of EVA1A:
Generating mutation plots.
Highly correlated genes for EVA1A:
Showing top 20/569 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EVA1A | CD276 | 0.708396 | 4 | 0 | 4 |
| EVA1A | MMP15 | 0.687566 | 3 | 0 | 3 |
| EVA1A | PLOD3 | 0.679148 | 5 | 0 | 5 |
| EVA1A | INHBA | 0.67828 | 6 | 0 | 5 |
| EVA1A | MFAP2 | 0.678269 | 5 | 0 | 5 |
| EVA1A | LAMA3 | 0.675577 | 4 | 0 | 4 |
| EVA1A | SDCCAG8 | 0.675539 | 3 | 0 | 3 |
| EVA1A | ACOT9 | 0.671261 | 4 | 0 | 4 |
| EVA1A | KIF2C | 0.668643 | 5 | 0 | 5 |
| EVA1A | AGRN | 0.668443 | 4 | 0 | 4 |
| EVA1A | MSN | 0.668417 | 4 | 0 | 4 |
| EVA1A | PCNA | 0.665321 | 4 | 0 | 3 |
| EVA1A | PON3 | 0.66384 | 3 | 0 | 3 |
| EVA1A | PTK7 | 0.663513 | 5 | 0 | 5 |
| EVA1A | COL5A1 | 0.663491 | 6 | 0 | 5 |
| EVA1A | PSMB2 | 0.662616 | 5 | 0 | 5 |
| EVA1A | VAV2 | 0.660255 | 4 | 0 | 4 |
| EVA1A | HOXD11 | 0.658783 | 4 | 0 | 4 |
| EVA1A | PDE7A | 0.658438 | 4 | 0 | 4 |
| EVA1A | CHCHD2 | 0.655269 | 3 | 0 | 3 |
For details and further investigation, click here