| Full name: major histocompatibility complex, class I, C | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6p21.33 | ||
| Entrez ID: 3107 | HGNC ID: HGNC:4933 | Ensembl Gene: ENSG00000204525 | OMIM ID: 142840 |
| Drug and gene relationship at DGIdb | |||
HLA-C involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04612 | Antigen processing and presentation | |
| hsa04650 | Natural killer cell mediated cytotoxicity | |
| hsa05166 | HTLV-I infection | |
| hsa05169 | Epstein-Barr virus infection |
Expression of HLA-C:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HLA-C | 3107 | 208812_x_at | 0.4183 | 0.4281 | |
| GSE20347 | HLA-C | 3107 | 208812_x_at | 0.4485 | 0.0683 | |
| GSE23400 | HLA-C | 3107 | 208812_x_at | 0.4765 | 0.0000 | |
| GSE26886 | HLA-C | 3107 | 208812_x_at | 0.1776 | 0.5926 | |
| GSE29001 | HLA-C | 3107 | 208812_x_at | 0.6477 | 0.0128 | |
| GSE38129 | HLA-C | 3107 | 208812_x_at | 0.3864 | 0.0329 | |
| GSE45670 | HLA-C | 3107 | 208812_x_at | 0.5579 | 0.0282 | |
| GSE53622 | HLA-C | 3107 | 64795 | 1.0348 | 0.0000 | |
| GSE53624 | HLA-C | 3107 | 64795 | 0.9787 | 0.0000 | |
| GSE63941 | HLA-C | 3107 | 208812_x_at | 0.6353 | 0.4120 | |
| GSE77861 | HLA-C | 3107 | 208812_x_at | 0.4266 | 0.0650 | |
| GSE97050 | HLA-C | 3107 | A_33_P3424803 | 1.0294 | 0.0905 | |
| SRP007169 | HLA-C | 3107 | RNAseq | 0.7246 | 0.1712 | |
| SRP008496 | HLA-C | 3107 | RNAseq | 0.4431 | 0.2453 | |
| SRP064894 | HLA-C | 3107 | RNAseq | 1.0962 | 0.0011 | |
| SRP133303 | HLA-C | 3107 | RNAseq | 0.8380 | 0.1380 | |
| SRP159526 | HLA-C | 3107 | RNAseq | 0.6114 | 0.5092 | |
| SRP193095 | HLA-C | 3107 | RNAseq | 0.7508 | 0.0111 | |
| SRP219564 | HLA-C | 3107 | RNAseq | 1.0190 | 0.3525 | |
| TCGA | HLA-C | 3107 | RNAseq | 0.1457 | 0.0416 |
Upregulated datasets: 2; Downregulated datasets: 0.
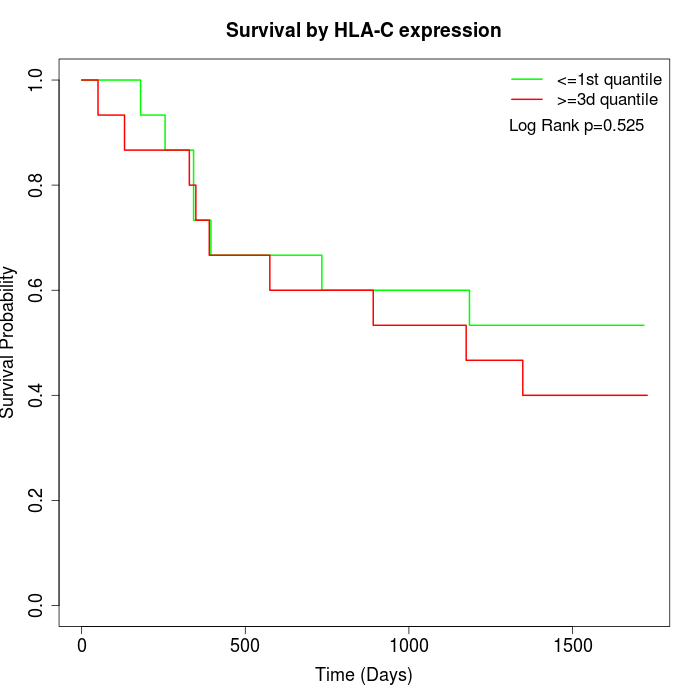
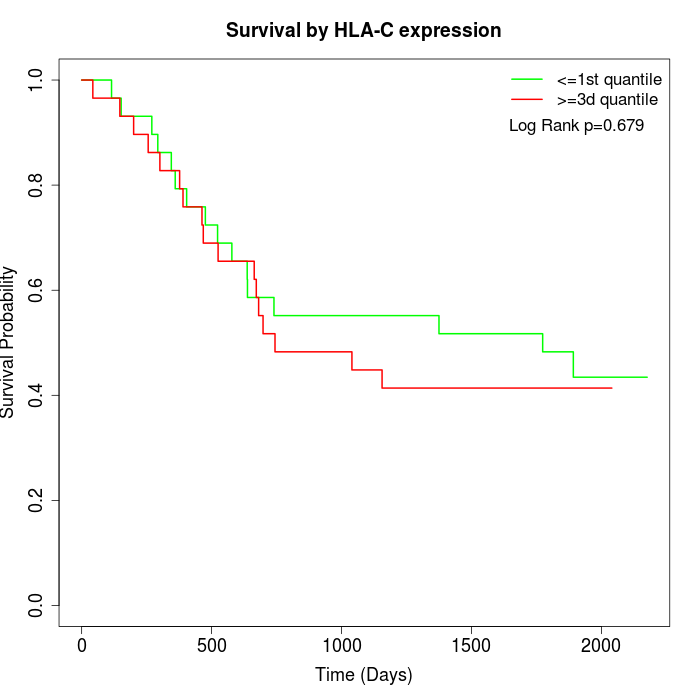
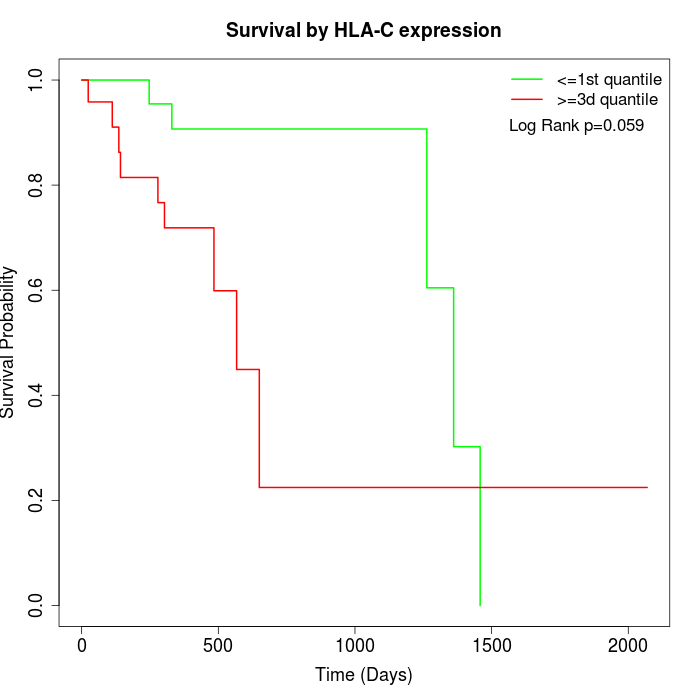
Survival by HLA-C expression:
Note: Click image to view full size file.
Copy number change of HLA-C:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HLA-C | 3107 | 5 | 1 | 24 | |
| GSE20123 | HLA-C | 3107 | 5 | 1 | 24 | |
| GSE43470 | HLA-C | 3107 | 5 | 1 | 37 | |
| GSE46452 | HLA-C | 3107 | 1 | 10 | 48 | |
| GSE47630 | HLA-C | 3107 | 7 | 3 | 30 | |
| GSE54993 | HLA-C | 3107 | 2 | 1 | 67 | |
| GSE54994 | HLA-C | 3107 | 11 | 4 | 38 | |
| GSE60625 | HLA-C | 3107 | 0 | 1 | 10 | |
| GSE74703 | HLA-C | 3107 | 5 | 0 | 31 | |
| GSE74704 | HLA-C | 3107 | 2 | 0 | 18 |
Total number of gains: 43; Total number of losses: 22; Total Number of normals: 327.
Somatic mutations of HLA-C:
Generating mutation plots.
Highly correlated genes for HLA-C:
Showing top 20/537 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HLA-C | HLA-B | 0.941252 | 13 | 0 | 13 |
| HLA-C | HLA-A | 0.857538 | 13 | 0 | 13 |
| HLA-C | HLA-F | 0.85672 | 13 | 0 | 13 |
| HLA-C | HLA-G | 0.829265 | 13 | 0 | 13 |
| HLA-C | PSMB9 | 0.75734 | 13 | 0 | 13 |
| HLA-C | UBE2L6 | 0.737036 | 13 | 0 | 12 |
| HLA-C | B2M | 0.735678 | 12 | 0 | 12 |
| HLA-C | TAP1 | 0.693664 | 11 | 0 | 10 |
| HLA-C | PSMB8 | 0.692708 | 13 | 0 | 13 |
| HLA-C | RNF166 | 0.686517 | 3 | 0 | 3 |
| HLA-C | BTN3A3 | 0.657577 | 10 | 0 | 8 |
| HLA-C | STAT1 | 0.654237 | 11 | 0 | 10 |
| HLA-C | EPSTI1 | 0.651329 | 7 | 0 | 6 |
| HLA-C | TAPBP | 0.648718 | 13 | 0 | 9 |
| HLA-C | PARP14 | 0.645421 | 9 | 0 | 7 |
| HLA-C | CXCL9 | 0.645025 | 12 | 0 | 10 |
| HLA-C | IFIH1 | 0.644648 | 9 | 0 | 8 |
| HLA-C | TMC8 | 0.644639 | 4 | 0 | 4 |
| HLA-C | UHMK1 | 0.643986 | 5 | 0 | 5 |
| HLA-C | CD80 | 0.643666 | 3 | 0 | 3 |
For details and further investigation, click here