| Full name: interferon gamma | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12q15 | ||
| Entrez ID: 3458 | HGNC ID: HGNC:5438 | Ensembl Gene: ENSG00000111537 | OMIM ID: 147570 |
| Drug and gene relationship at DGIdb | |||
IFNG involved pathways:
Expression of IFNG:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | IFNG | 3458 | 210354_at | 0.0526 | 0.9687 | |
| GSE20347 | IFNG | 3458 | 210354_at | 0.2384 | 0.1561 | |
| GSE23400 | IFNG | 3458 | 210354_at | -0.0031 | 0.9447 | |
| GSE26886 | IFNG | 3458 | 210354_at | -0.0465 | 0.8691 | |
| GSE29001 | IFNG | 3458 | 210354_at | 0.0845 | 0.6534 | |
| GSE38129 | IFNG | 3458 | 210354_at | 0.1487 | 0.2101 | |
| GSE45670 | IFNG | 3458 | 210354_at | 0.3619 | 0.1796 | |
| GSE53622 | IFNG | 3458 | 49485 | 0.5157 | 0.0056 | |
| GSE53624 | IFNG | 3458 | 49485 | 0.5517 | 0.0021 | |
| GSE63941 | IFNG | 3458 | 210354_at | 0.1861 | 0.1466 | |
| GSE77861 | IFNG | 3458 | 210354_at | -0.1507 | 0.0854 | |
| GSE97050 | IFNG | 3458 | A_23_P151294 | 0.1910 | 0.3801 | |
| TCGA | IFNG | 3458 | RNAseq | 2.9879 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
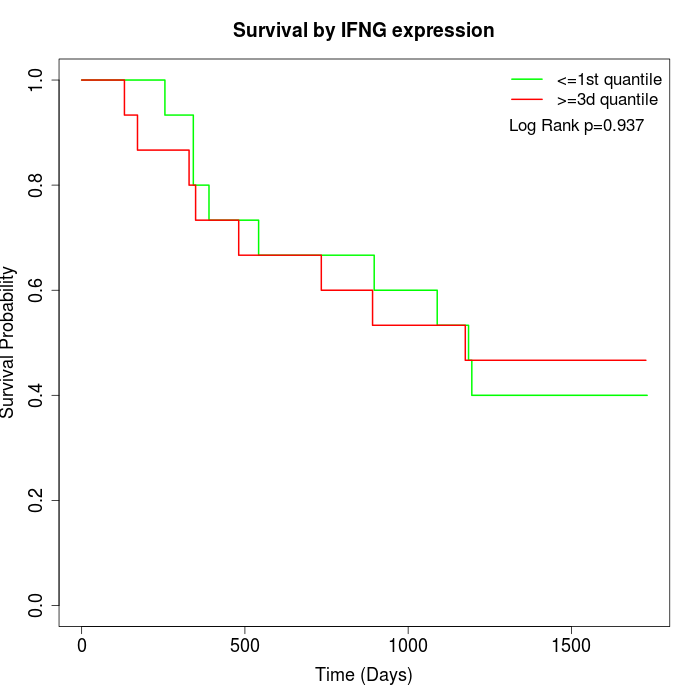
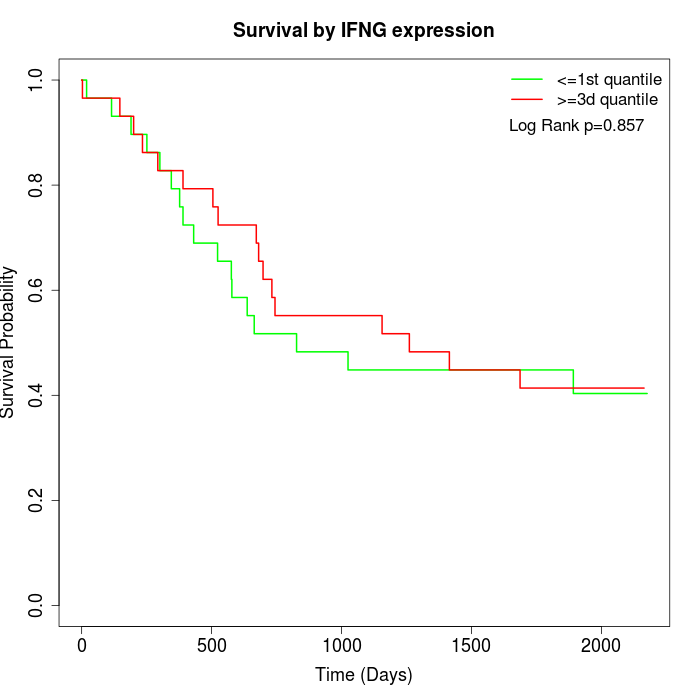
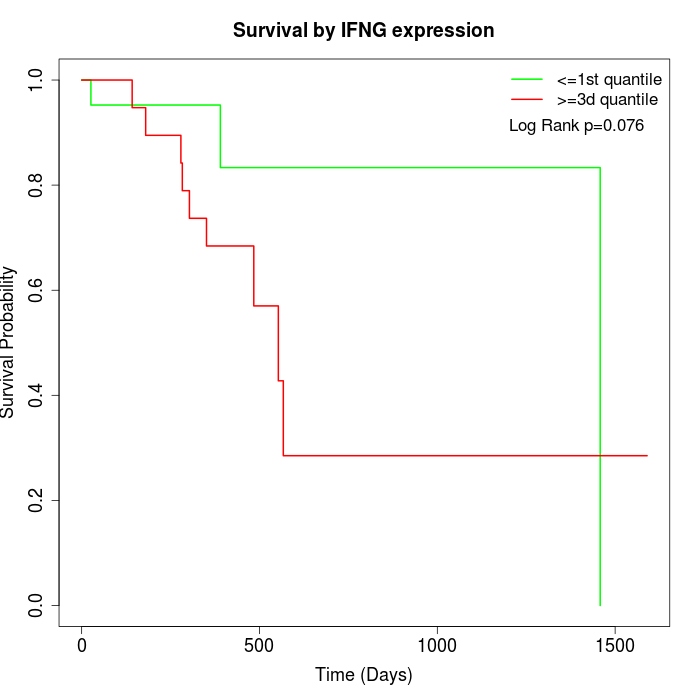
Survival by IFNG expression:
Note: Click image to view full size file.
Copy number change of IFNG:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | IFNG | 3458 | 6 | 0 | 24 | |
| GSE20123 | IFNG | 3458 | 6 | 0 | 24 | |
| GSE43470 | IFNG | 3458 | 5 | 0 | 38 | |
| GSE46452 | IFNG | 3458 | 13 | 0 | 46 | |
| GSE47630 | IFNG | 3458 | 10 | 1 | 29 | |
| GSE54993 | IFNG | 3458 | 0 | 8 | 62 | |
| GSE54994 | IFNG | 3458 | 7 | 1 | 45 | |
| GSE60625 | IFNG | 3458 | 0 | 0 | 11 | |
| GSE74703 | IFNG | 3458 | 5 | 0 | 31 | |
| GSE74704 | IFNG | 3458 | 5 | 0 | 15 | |
| TCGA | IFNG | 3458 | 24 | 9 | 63 |
Total number of gains: 81; Total number of losses: 19; Total Number of normals: 388.
Somatic mutations of IFNG:
Generating mutation plots.
Highly correlated genes for IFNG:
Showing top 20/115 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| IFNG | GDF5 | 0.678972 | 3 | 0 | 3 |
| IFNG | KLRC4 | 0.658022 | 4 | 0 | 3 |
| IFNG | PRF1 | 0.649269 | 8 | 0 | 7 |
| IFNG | BATF2 | 0.623803 | 6 | 0 | 5 |
| IFNG | CXCL11 | 0.622193 | 11 | 0 | 8 |
| IFNG | AKAP5 | 0.617828 | 7 | 0 | 5 |
| IFNG | AKAP8L | 0.616936 | 3 | 0 | 3 |
| IFNG | TLR4 | 0.614152 | 3 | 0 | 3 |
| IFNG | GBP5 | 0.613156 | 6 | 0 | 4 |
| IFNG | LAG3 | 0.606508 | 10 | 0 | 7 |
| IFNG | KLRD1 | 0.599848 | 8 | 0 | 7 |
| IFNG | CHRNA9 | 0.595883 | 4 | 0 | 3 |
| IFNG | AHI1 | 0.594849 | 4 | 0 | 3 |
| IFNG | EPSTI1 | 0.591897 | 7 | 0 | 6 |
| IFNG | GBP4 | 0.591494 | 8 | 0 | 6 |
| IFNG | NKG7 | 0.591321 | 12 | 0 | 8 |
| IFNG | GZMB | 0.588807 | 11 | 0 | 9 |
| IFNG | LRGUK | 0.587229 | 4 | 0 | 3 |
| IFNG | SAMD3 | 0.581086 | 6 | 0 | 5 |
| IFNG | CACNG3 | 0.576545 | 4 | 0 | 3 |
For details and further investigation, click here