| Full name: lipopolysaccharide induced TNF factor | Alias Symbol: PIG7|SIMPLE|FLJ38636|TP53I7 | ||
| Type: protein-coding gene | Cytoband: 16p13.13 | ||
| Entrez ID: 9516 | HGNC ID: HGNC:16841 | Ensembl Gene: ENSG00000189067 | OMIM ID: 603795 |
| Drug and gene relationship at DGIdb | |||
Expression of LITAF:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LITAF | 9516 | 200704_at | 0.0794 | 0.9177 | |
| GSE20347 | LITAF | 9516 | 200704_at | 0.8783 | 0.0001 | |
| GSE23400 | LITAF | 9516 | 200704_at | 0.7090 | 0.0000 | |
| GSE26886 | LITAF | 9516 | 200704_at | 0.7258 | 0.0435 | |
| GSE29001 | LITAF | 9516 | 200704_at | 0.9863 | 0.0005 | |
| GSE38129 | LITAF | 9516 | 200704_at | 0.7790 | 0.0002 | |
| GSE45670 | LITAF | 9516 | 200704_at | 0.3678 | 0.1084 | |
| GSE53622 | LITAF | 9516 | 115156 | 0.6145 | 0.0000 | |
| GSE53624 | LITAF | 9516 | 115156 | 0.4131 | 0.0000 | |
| GSE63941 | LITAF | 9516 | 200706_s_at | 0.6558 | 0.2268 | |
| GSE77861 | LITAF | 9516 | 200704_at | 0.9357 | 0.0082 | |
| GSE97050 | LITAF | 9516 | A_23_P3532 | 0.4648 | 0.1646 | |
| SRP007169 | LITAF | 9516 | RNAseq | 1.5378 | 0.0002 | |
| SRP008496 | LITAF | 9516 | RNAseq | 1.4311 | 0.0000 | |
| SRP064894 | LITAF | 9516 | RNAseq | 0.0359 | 0.8796 | |
| SRP133303 | LITAF | 9516 | RNAseq | 0.9165 | 0.0000 | |
| SRP159526 | LITAF | 9516 | RNAseq | 0.7121 | 0.0111 | |
| SRP193095 | LITAF | 9516 | RNAseq | 0.9132 | 0.0000 | |
| SRP219564 | LITAF | 9516 | RNAseq | 0.7333 | 0.0060 | |
| TCGA | LITAF | 9516 | RNAseq | 0.0632 | 0.2009 |
Upregulated datasets: 2; Downregulated datasets: 0.
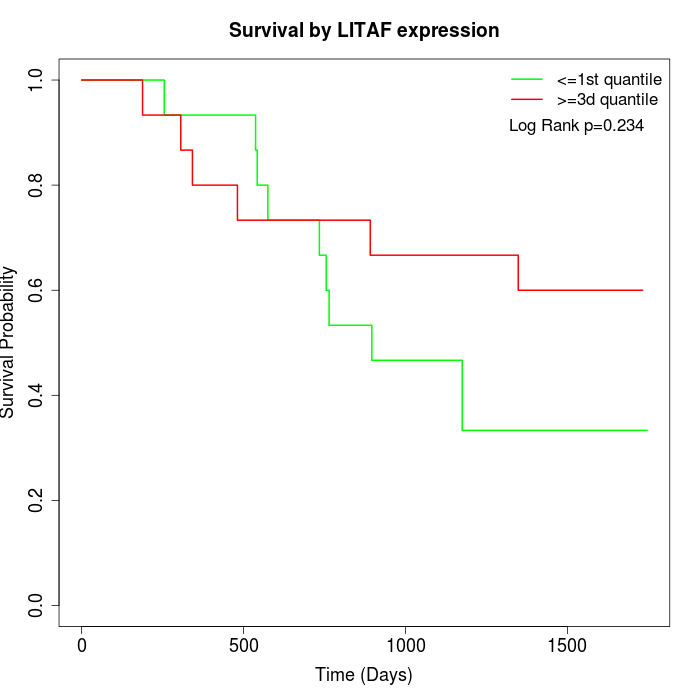
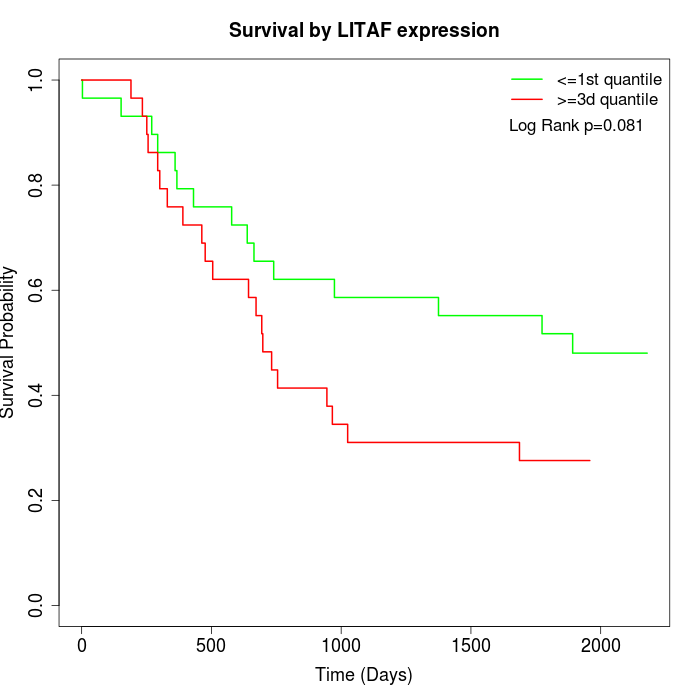
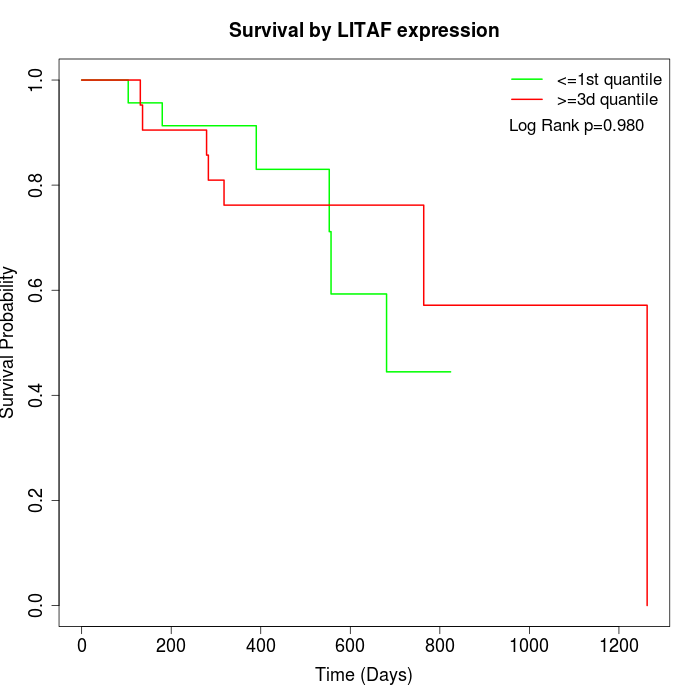
Survival by LITAF expression:
Note: Click image to view full size file.
Copy number change of LITAF:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LITAF | 9516 | 5 | 4 | 21 | |
| GSE20123 | LITAF | 9516 | 5 | 3 | 22 | |
| GSE43470 | LITAF | 9516 | 3 | 3 | 37 | |
| GSE46452 | LITAF | 9516 | 38 | 1 | 20 | |
| GSE47630 | LITAF | 9516 | 14 | 6 | 20 | |
| GSE54993 | LITAF | 9516 | 3 | 5 | 62 | |
| GSE54994 | LITAF | 9516 | 5 | 9 | 39 | |
| GSE60625 | LITAF | 9516 | 4 | 0 | 7 | |
| GSE74703 | LITAF | 9516 | 3 | 2 | 31 | |
| GSE74704 | LITAF | 9516 | 2 | 1 | 17 | |
| TCGA | LITAF | 9516 | 21 | 12 | 63 |
Total number of gains: 103; Total number of losses: 46; Total Number of normals: 339.
Somatic mutations of LITAF:
Generating mutation plots.
Highly correlated genes for LITAF:
Showing top 20/1277 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LITAF | PREX1 | 0.820176 | 3 | 0 | 3 |
| LITAF | TAGAP | 0.802092 | 3 | 0 | 3 |
| LITAF | FCGR3A | 0.742594 | 3 | 0 | 3 |
| LITAF | NUB1 | 0.74208 | 3 | 0 | 3 |
| LITAF | VPS16 | 0.737444 | 4 | 0 | 3 |
| LITAF | GCLC | 0.736948 | 3 | 0 | 3 |
| LITAF | NSMCE4A | 0.72698 | 3 | 0 | 3 |
| LITAF | CERKL | 0.713814 | 3 | 0 | 3 |
| LITAF | GMEB2 | 0.711199 | 3 | 0 | 3 |
| LITAF | LIN9 | 0.702591 | 3 | 0 | 3 |
| LITAF | MRPL47 | 0.701012 | 4 | 0 | 4 |
| LITAF | C9orf64 | 0.699215 | 3 | 0 | 3 |
| LITAF | TNFRSF1A | 0.697455 | 3 | 0 | 3 |
| LITAF | TMEM60 | 0.696238 | 3 | 0 | 3 |
| LITAF | HPS3 | 0.694612 | 4 | 0 | 3 |
| LITAF | DPYSL2 | 0.692609 | 7 | 0 | 6 |
| LITAF | PIGX | 0.691432 | 5 | 0 | 3 |
| LITAF | EDC4 | 0.686268 | 5 | 0 | 5 |
| LITAF | VRK2 | 0.686056 | 3 | 0 | 3 |
| LITAF | NFE2L1 | 0.685221 | 8 | 0 | 6 |
For details and further investigation, click here