| Full name: mitogen-activated protein kinase kinase kinase kinase 4 | Alias Symbol: HGK|NIK|FLH21957 | ||
| Type: protein-coding gene | Cytoband: 2q11.2 | ||
| Entrez ID: 9448 | HGNC ID: HGNC:6866 | Ensembl Gene: ENSG00000071054 | OMIM ID: 604666 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
MAP4K4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway |
Expression of MAP4K4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP4K4 | 9448 | 206571_s_at | 0.8550 | 0.0827 | |
| GSE20347 | MAP4K4 | 9448 | 206571_s_at | -0.1127 | 0.5791 | |
| GSE23400 | MAP4K4 | 9448 | 206571_s_at | 0.2891 | 0.0016 | |
| GSE26886 | MAP4K4 | 9448 | 206571_s_at | -0.8497 | 0.0073 | |
| GSE29001 | MAP4K4 | 9448 | 206571_s_at | -0.2253 | 0.5411 | |
| GSE38129 | MAP4K4 | 9448 | 206571_s_at | 0.3012 | 0.0467 | |
| GSE45670 | MAP4K4 | 9448 | 206571_s_at | 0.6428 | 0.0037 | |
| GSE53622 | MAP4K4 | 9448 | 40265 | 0.0170 | 0.8585 | |
| GSE53624 | MAP4K4 | 9448 | 40265 | 0.2671 | 0.0062 | |
| GSE63941 | MAP4K4 | 9448 | 206571_s_at | -1.8739 | 0.0040 | |
| GSE77861 | MAP4K4 | 9448 | 206571_s_at | 0.3364 | 0.2612 | |
| GSE97050 | MAP4K4 | 9448 | A_23_P90804 | 0.2380 | 0.2850 | |
| SRP007169 | MAP4K4 | 9448 | RNAseq | 0.6722 | 0.1285 | |
| SRP008496 | MAP4K4 | 9448 | RNAseq | 0.7353 | 0.0073 | |
| SRP064894 | MAP4K4 | 9448 | RNAseq | 0.4194 | 0.0091 | |
| SRP133303 | MAP4K4 | 9448 | RNAseq | 0.5775 | 0.0008 | |
| SRP159526 | MAP4K4 | 9448 | RNAseq | 0.2429 | 0.4484 | |
| SRP193095 | MAP4K4 | 9448 | RNAseq | 0.4436 | 0.0164 | |
| SRP219564 | MAP4K4 | 9448 | RNAseq | -0.0833 | 0.8298 | |
| TCGA | MAP4K4 | 9448 | RNAseq | 0.2121 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
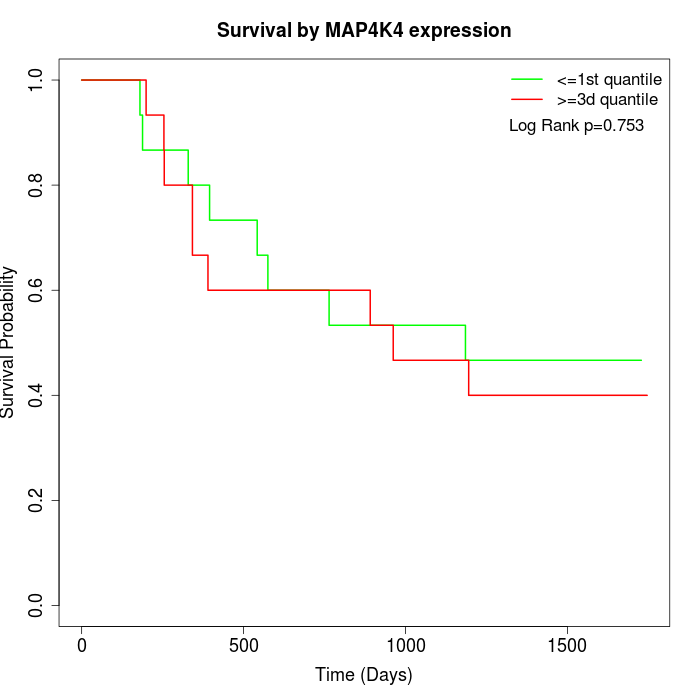
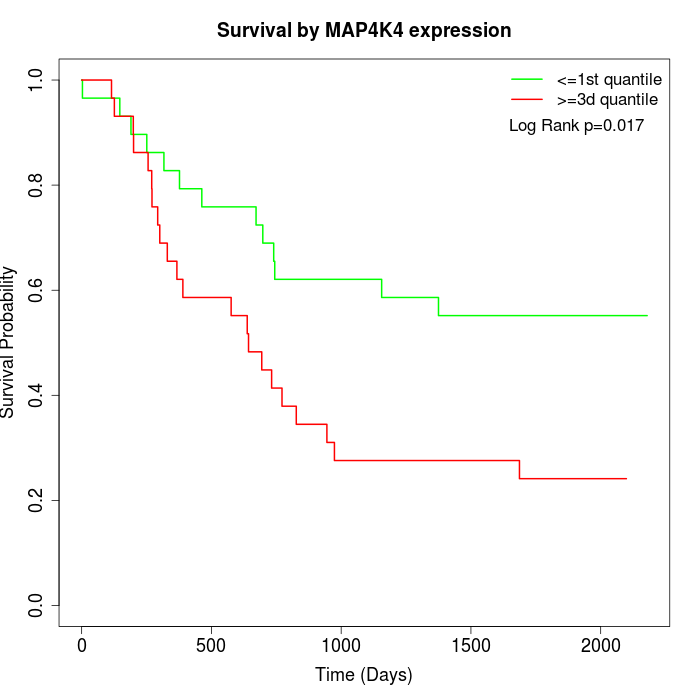
Survival by MAP4K4 expression:
Note: Click image to view full size file.
Copy number change of MAP4K4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP4K4 | 9448 | 5 | 2 | 23 | |
| GSE20123 | MAP4K4 | 9448 | 5 | 2 | 23 | |
| GSE43470 | MAP4K4 | 9448 | 5 | 1 | 37 | |
| GSE46452 | MAP4K4 | 9448 | 1 | 4 | 54 | |
| GSE47630 | MAP4K4 | 9448 | 7 | 0 | 33 | |
| GSE54993 | MAP4K4 | 9448 | 0 | 6 | 64 | |
| GSE54994 | MAP4K4 | 9448 | 11 | 0 | 42 | |
| GSE60625 | MAP4K4 | 9448 | 0 | 3 | 8 | |
| GSE74703 | MAP4K4 | 9448 | 5 | 1 | 30 | |
| GSE74704 | MAP4K4 | 9448 | 3 | 1 | 16 | |
| TCGA | MAP4K4 | 9448 | 35 | 6 | 55 |
Total number of gains: 77; Total number of losses: 26; Total Number of normals: 385.
Somatic mutations of MAP4K4:
Generating mutation plots.
Highly correlated genes for MAP4K4:
Showing top 20/512 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP4K4 | CIAO1 | 0.815046 | 3 | 0 | 3 |
| MAP4K4 | NUDT5 | 0.807759 | 3 | 0 | 3 |
| MAP4K4 | GPD2 | 0.784701 | 3 | 0 | 3 |
| MAP4K4 | MKNK1 | 0.776405 | 3 | 0 | 3 |
| MAP4K4 | ALG2 | 0.76252 | 3 | 0 | 3 |
| MAP4K4 | DCAF13 | 0.760222 | 3 | 0 | 3 |
| MAP4K4 | TRADD | 0.7564 | 3 | 0 | 3 |
| MAP4K4 | MASTL | 0.753022 | 3 | 0 | 3 |
| MAP4K4 | STC2 | 0.750786 | 4 | 0 | 4 |
| MAP4K4 | BDKRB1 | 0.749923 | 3 | 0 | 3 |
| MAP4K4 | TPM3 | 0.747174 | 4 | 0 | 4 |
| MAP4K4 | TERF2IP | 0.740508 | 3 | 0 | 3 |
| MAP4K4 | C11orf24 | 0.739638 | 4 | 0 | 4 |
| MAP4K4 | CDC42SE1 | 0.738759 | 3 | 0 | 3 |
| MAP4K4 | IRF2BPL | 0.737466 | 3 | 0 | 3 |
| MAP4K4 | TP53I11 | 0.736556 | 3 | 0 | 3 |
| MAP4K4 | IFFO2 | 0.736481 | 3 | 0 | 3 |
| MAP4K4 | ERLEC1 | 0.736409 | 3 | 0 | 3 |
| MAP4K4 | ATG7 | 0.736238 | 3 | 0 | 3 |
| MAP4K4 | BMP2K | 0.733493 | 3 | 0 | 3 |
For details and further investigation, click here