| Full name: pleckstrin homology and RUN domain containing M2 | Alias Symbol: KIAA0842 | ||
| Type: protein-coding gene | Cytoband: 1p36.21 | ||
| Entrez ID: 23207 | HGNC ID: HGNC:29131 | Ensembl Gene: ENSG00000116786 | OMIM ID: 609613 |
| Drug and gene relationship at DGIdb | |||
PLEKHM2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05132 | Salmonella infection |
Expression of PLEKHM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLEKHM2 | 23207 | 212146_at | 0.3033 | 0.4787 | |
| GSE20347 | PLEKHM2 | 23207 | 212146_at | -0.0958 | 0.4620 | |
| GSE23400 | PLEKHM2 | 23207 | 212146_at | -0.0257 | 0.5999 | |
| GSE26886 | PLEKHM2 | 23207 | 212146_at | -0.2284 | 0.3080 | |
| GSE29001 | PLEKHM2 | 23207 | 212146_at | -0.1944 | 0.3568 | |
| GSE38129 | PLEKHM2 | 23207 | 212146_at | -0.0329 | 0.8184 | |
| GSE45670 | PLEKHM2 | 23207 | 212146_at | 0.0969 | 0.4627 | |
| GSE53622 | PLEKHM2 | 23207 | 1342 | 0.0600 | 0.4385 | |
| GSE53624 | PLEKHM2 | 23207 | 1342 | 0.0248 | 0.7179 | |
| GSE63941 | PLEKHM2 | 23207 | 212146_at | -0.4958 | 0.1593 | |
| GSE77861 | PLEKHM2 | 23207 | 212146_at | -0.0295 | 0.8959 | |
| GSE97050 | PLEKHM2 | 23207 | A_23_P351342 | 0.4011 | 0.1660 | |
| SRP007169 | PLEKHM2 | 23207 | RNAseq | -1.1768 | 0.0122 | |
| SRP008496 | PLEKHM2 | 23207 | RNAseq | -1.0245 | 0.0001 | |
| SRP064894 | PLEKHM2 | 23207 | RNAseq | 0.0483 | 0.6948 | |
| SRP133303 | PLEKHM2 | 23207 | RNAseq | -0.0256 | 0.8894 | |
| SRP159526 | PLEKHM2 | 23207 | RNAseq | -0.3459 | 0.0935 | |
| SRP193095 | PLEKHM2 | 23207 | RNAseq | 0.0190 | 0.8910 | |
| SRP219564 | PLEKHM2 | 23207 | RNAseq | -0.4456 | 0.0077 | |
| TCGA | PLEKHM2 | 23207 | RNAseq | 0.0370 | 0.3975 |
Upregulated datasets: 0; Downregulated datasets: 2.
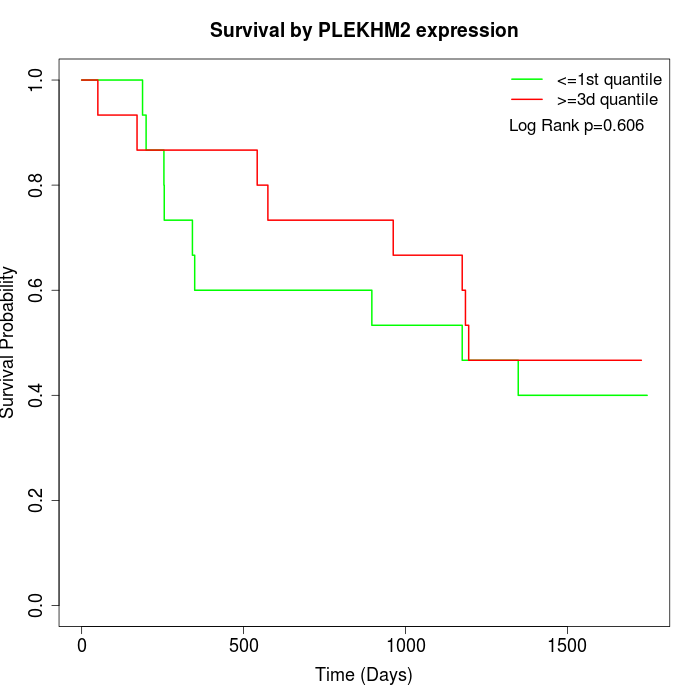
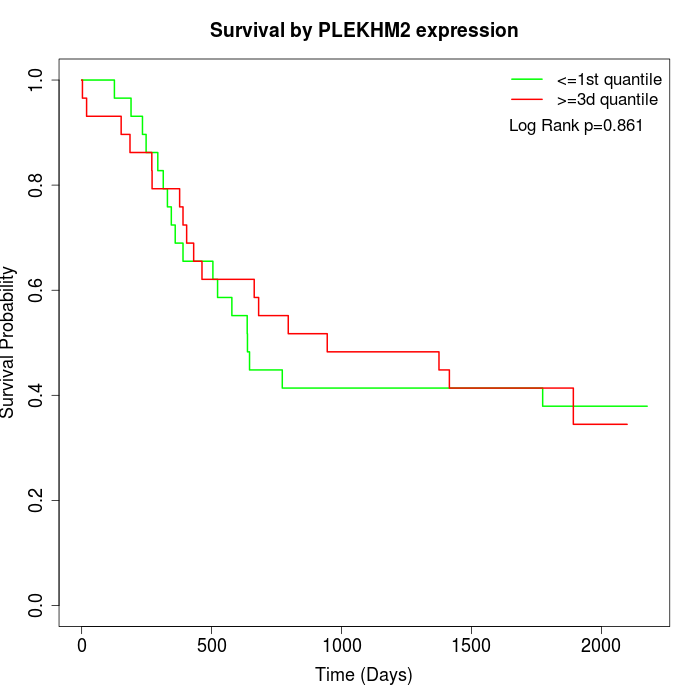
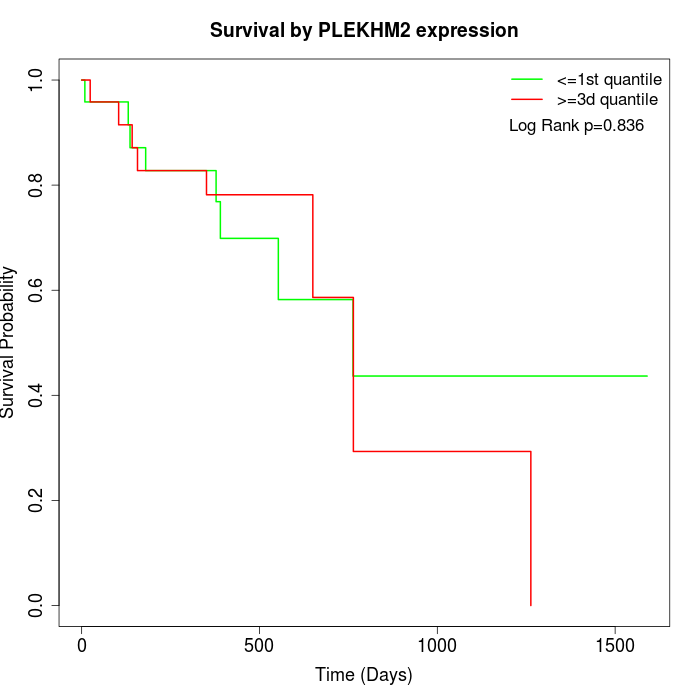
Survival by PLEKHM2 expression:
Note: Click image to view full size file.
Copy number change of PLEKHM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PLEKHM2 | 23207 | 1 | 5 | 24 | |
| GSE20123 | PLEKHM2 | 23207 | 1 | 4 | 25 | |
| GSE43470 | PLEKHM2 | 23207 | 1 | 8 | 34 | |
| GSE46452 | PLEKHM2 | 23207 | 7 | 1 | 51 | |
| GSE47630 | PLEKHM2 | 23207 | 8 | 3 | 29 | |
| GSE54993 | PLEKHM2 | 23207 | 3 | 1 | 66 | |
| GSE54994 | PLEKHM2 | 23207 | 10 | 8 | 35 | |
| GSE60625 | PLEKHM2 | 23207 | 0 | 0 | 11 | |
| GSE74703 | PLEKHM2 | 23207 | 1 | 5 | 30 | |
| GSE74704 | PLEKHM2 | 23207 | 1 | 0 | 19 | |
| TCGA | PLEKHM2 | 23207 | 12 | 23 | 61 |
Total number of gains: 45; Total number of losses: 58; Total Number of normals: 385.
Somatic mutations of PLEKHM2:
Generating mutation plots.
Highly correlated genes for PLEKHM2:
Showing top 20/296 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLEKHM2 | BCAR3 | 0.808372 | 3 | 0 | 3 |
| PLEKHM2 | YIPF3 | 0.790535 | 3 | 0 | 3 |
| PLEKHM2 | IFT80 | 0.787853 | 3 | 0 | 3 |
| PLEKHM2 | ZNHIT1 | 0.785719 | 3 | 0 | 3 |
| PLEKHM2 | METAP1 | 0.74966 | 3 | 0 | 3 |
| PLEKHM2 | PUS10 | 0.749294 | 3 | 0 | 3 |
| PLEKHM2 | TMEM41A | 0.745162 | 3 | 0 | 3 |
| PLEKHM2 | GLE1 | 0.744449 | 3 | 0 | 3 |
| PLEKHM2 | E2F3 | 0.729652 | 3 | 0 | 3 |
| PLEKHM2 | TRUB2 | 0.726356 | 3 | 0 | 3 |
| PLEKHM2 | POLRMT | 0.718572 | 4 | 0 | 4 |
| PLEKHM2 | LRP11 | 0.710428 | 3 | 0 | 3 |
| PLEKHM2 | HGS | 0.706361 | 6 | 0 | 5 |
| PLEKHM2 | USP42 | 0.705189 | 3 | 0 | 3 |
| PLEKHM2 | MIF | 0.696238 | 3 | 0 | 3 |
| PLEKHM2 | INTS10 | 0.692482 | 3 | 0 | 3 |
| PLEKHM2 | RABEP2 | 0.691516 | 5 | 0 | 4 |
| PLEKHM2 | RFXANK | 0.686427 | 4 | 0 | 3 |
| PLEKHM2 | MRPL37 | 0.684192 | 4 | 0 | 3 |
| PLEKHM2 | NAA60 | 0.683223 | 4 | 0 | 3 |
For details and further investigation, click here