| Full name: plastin 3 | Alias Symbol: T-plastin | ||
| Type: protein-coding gene | Cytoband: Xq23 | ||
| Entrez ID: 5358 | HGNC ID: HGNC:9091 | Ensembl Gene: ENSG00000102024 | OMIM ID: 300131 |
| Drug and gene relationship at DGIdb | |||
Expression of PLS3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLS3 | 5358 | 201215_at | -0.5250 | 0.4065 | |
| GSE20347 | PLS3 | 5358 | 201215_at | -0.5063 | 0.0046 | |
| GSE23400 | PLS3 | 5358 | 201215_at | 0.0461 | 0.6638 | |
| GSE26886 | PLS3 | 5358 | 201215_at | -0.5368 | 0.0028 | |
| GSE29001 | PLS3 | 5358 | 201215_at | -0.1897 | 0.6167 | |
| GSE38129 | PLS3 | 5358 | 201215_at | -0.1377 | 0.4611 | |
| GSE45670 | PLS3 | 5358 | 201215_at | 0.0031 | 0.9844 | |
| GSE53622 | PLS3 | 5358 | 7880 | 0.0862 | 0.5110 | |
| GSE53624 | PLS3 | 5358 | 144841 | -0.2672 | 0.0000 | |
| GSE63941 | PLS3 | 5358 | 201215_at | 0.0906 | 0.8021 | |
| GSE77861 | PLS3 | 5358 | 201215_at | -0.2035 | 0.3735 | |
| GSE97050 | PLS3 | 5358 | A_23_P250607 | 0.3310 | 0.5820 | |
| SRP007169 | PLS3 | 5358 | RNAseq | -1.0729 | 0.0107 | |
| SRP008496 | PLS3 | 5358 | RNAseq | -0.9871 | 0.0004 | |
| SRP064894 | PLS3 | 5358 | RNAseq | -0.2509 | 0.3125 | |
| SRP133303 | PLS3 | 5358 | RNAseq | 0.2292 | 0.1923 | |
| SRP159526 | PLS3 | 5358 | RNAseq | -0.1291 | 0.8040 | |
| SRP193095 | PLS3 | 5358 | RNAseq | -0.5404 | 0.0544 | |
| SRP219564 | PLS3 | 5358 | RNAseq | -0.0477 | 0.9266 | |
| TCGA | PLS3 | 5358 | RNAseq | 0.2431 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 1.
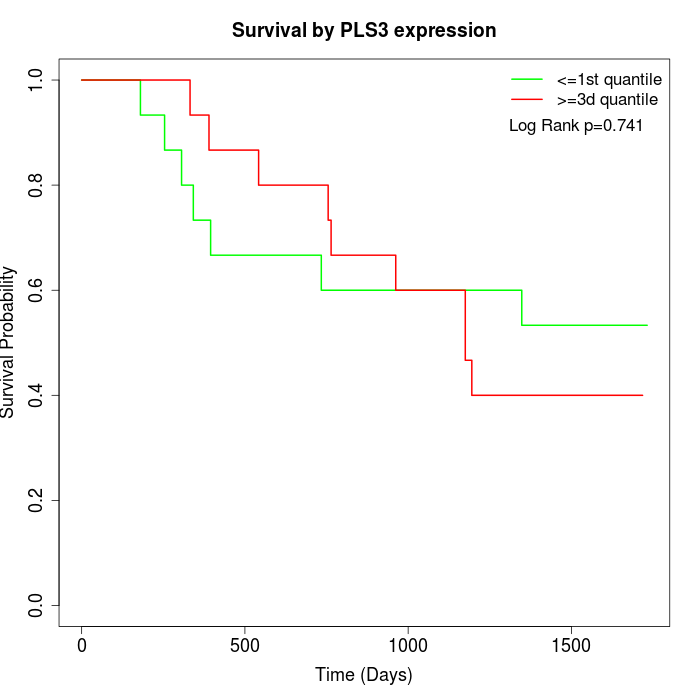
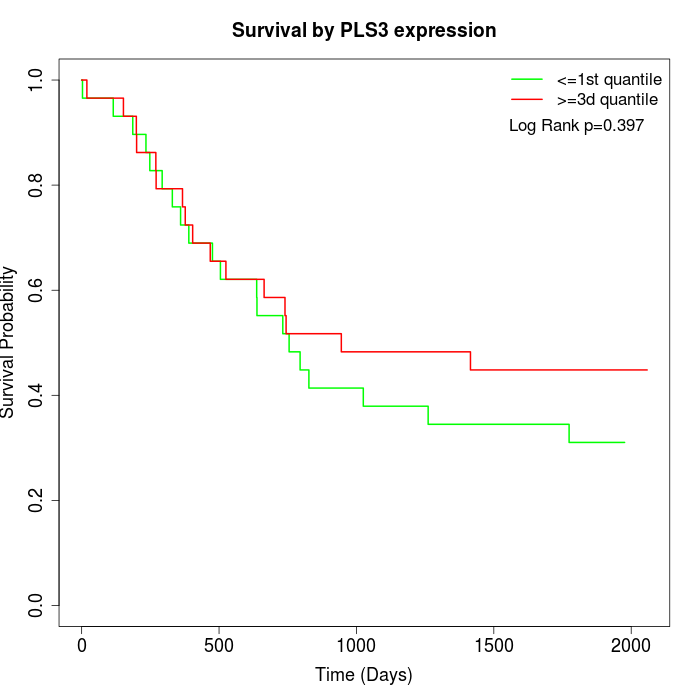
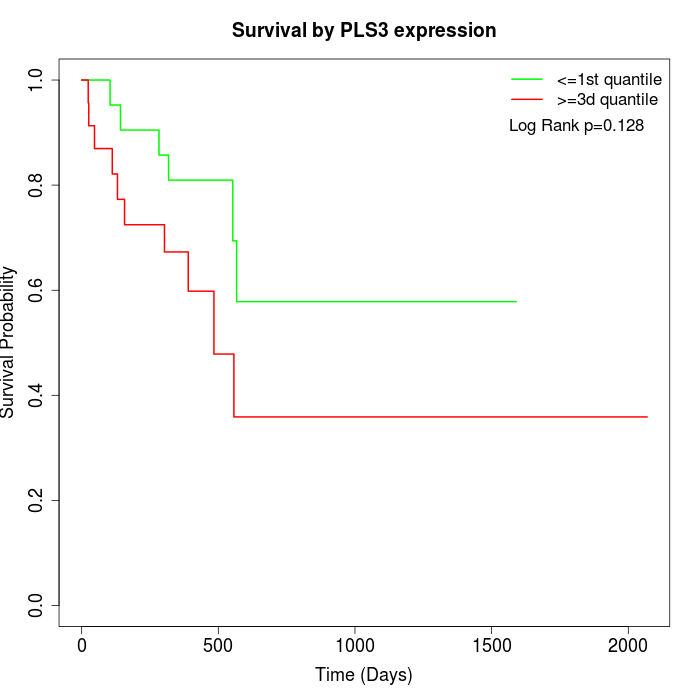
Survival by PLS3 expression:
Note: Click image to view full size file.
Copy number change of PLS3:
No record found for this gene.
Somatic mutations of PLS3:
Generating mutation plots.
Highly correlated genes for PLS3:
Showing top 20/612 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLS3 | GBP6 | 0.717854 | 3 | 0 | 3 |
| PLS3 | GJB6 | 0.714165 | 6 | 0 | 6 |
| PLS3 | NAMPT | 0.704306 | 6 | 0 | 6 |
| PLS3 | RNFT1 | 0.703998 | 3 | 0 | 3 |
| PLS3 | FAM126B | 0.702748 | 3 | 0 | 3 |
| PLS3 | GJB2 | 0.685917 | 4 | 0 | 3 |
| PLS3 | PFKFB2 | 0.67032 | 3 | 0 | 3 |
| PLS3 | CNEP1R1 | 0.664485 | 4 | 0 | 3 |
| PLS3 | RETSAT | 0.662153 | 3 | 0 | 3 |
| PLS3 | APPL2 | 0.659718 | 3 | 0 | 3 |
| PLS3 | KIF21A | 0.65912 | 4 | 0 | 4 |
| PLS3 | CMTR2 | 0.657831 | 4 | 0 | 3 |
| PLS3 | LEO1 | 0.65534 | 4 | 0 | 3 |
| PLS3 | DMKN | 0.653921 | 5 | 0 | 5 |
| PLS3 | SPNS2 | 0.653568 | 3 | 0 | 3 |
| PLS3 | SIAE | 0.649835 | 3 | 0 | 3 |
| PLS3 | PRKAR1A | 0.649299 | 4 | 0 | 3 |
| PLS3 | KRT6A | 0.647112 | 9 | 0 | 8 |
| PLS3 | FAR1 | 0.647054 | 5 | 0 | 4 |
| PLS3 | NDUFAF1 | 0.646341 | 5 | 0 | 4 |
For details and further investigation, click here