| Full name: proteasome activator subunit 2 | Alias Symbol: PA28beta | ||
| Type: protein-coding gene | Cytoband: 14q12 | ||
| Entrez ID: 5721 | HGNC ID: HGNC:9569 | Ensembl Gene: ENSG00000100911 | OMIM ID: 602161 |
| Drug and gene relationship at DGIdb | |||
PSME2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04612 | Antigen processing and presentation |
Expression of PSME2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | PSME2 | 5721 | 4850 | 0.4763 | 0.0000 | |
| GSE53624 | PSME2 | 5721 | 4850 | 0.7862 | 0.0000 | |
| GSE97050 | PSME2 | 5721 | A_23_P65427 | 0.9307 | 0.0866 | |
| SRP007169 | PSME2 | 5721 | RNAseq | 0.5208 | 0.2454 | |
| SRP008496 | PSME2 | 5721 | RNAseq | 0.0937 | 0.7510 | |
| SRP064894 | PSME2 | 5721 | RNAseq | 1.0514 | 0.0000 | |
| SRP133303 | PSME2 | 5721 | RNAseq | 0.8682 | 0.0000 | |
| SRP159526 | PSME2 | 5721 | RNAseq | 0.3266 | 0.3482 | |
| SRP193095 | PSME2 | 5721 | RNAseq | 0.2499 | 0.1384 | |
| SRP219564 | PSME2 | 5721 | RNAseq | 0.9503 | 0.0295 | |
| TCGA | PSME2 | 5721 | RNAseq | 0.3432 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
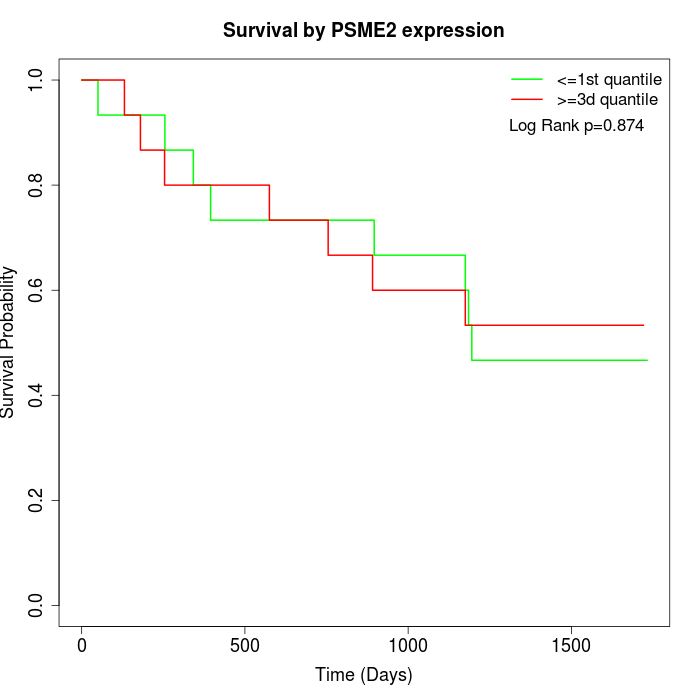
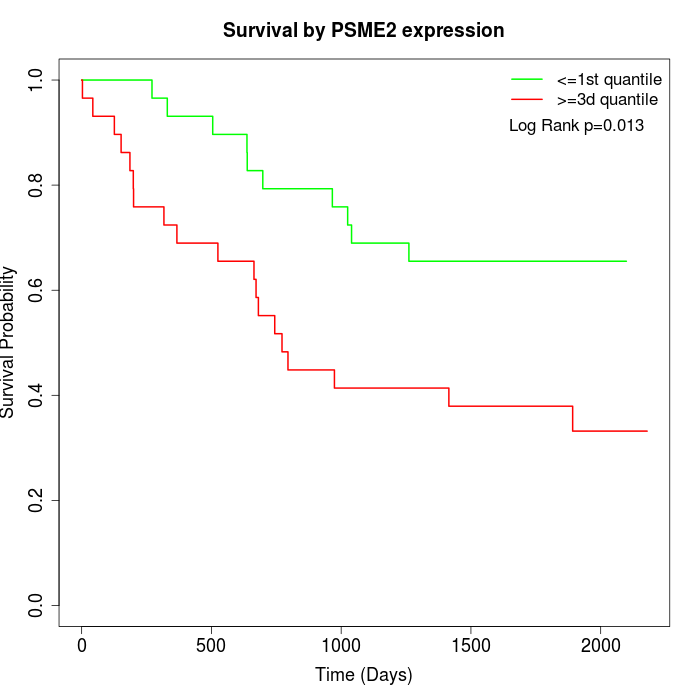
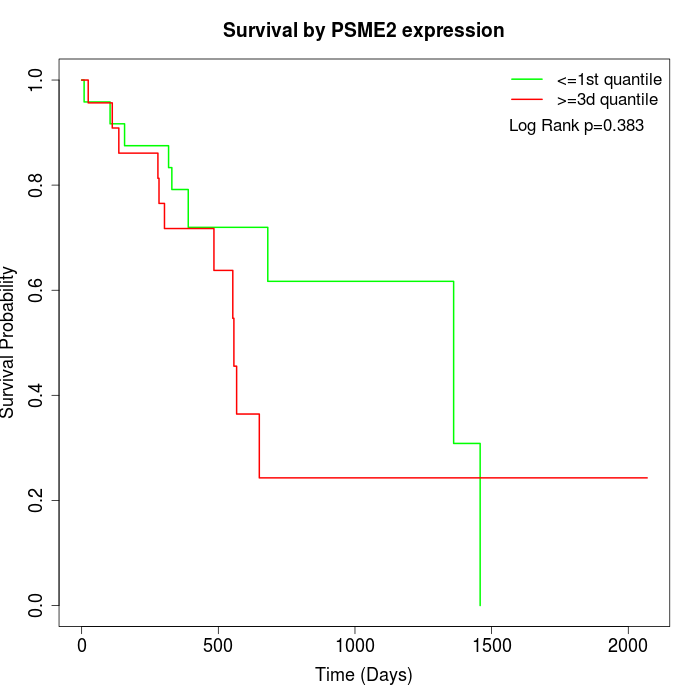
Survival by PSME2 expression:
Note: Click image to view full size file.
Copy number change of PSME2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PSME2 | 5721 | 8 | 3 | 19 | |
| GSE20123 | PSME2 | 5721 | 8 | 3 | 19 | |
| GSE43470 | PSME2 | 5721 | 8 | 2 | 33 | |
| GSE46452 | PSME2 | 5721 | 16 | 2 | 41 | |
| GSE47630 | PSME2 | 5721 | 11 | 10 | 19 | |
| GSE54993 | PSME2 | 5721 | 3 | 11 | 56 | |
| GSE54994 | PSME2 | 5721 | 18 | 5 | 30 | |
| GSE60625 | PSME2 | 5721 | 0 | 2 | 9 | |
| GSE74703 | PSME2 | 5721 | 7 | 2 | 27 | |
| GSE74704 | PSME2 | 5721 | 3 | 2 | 15 | |
| TCGA | PSME2 | 5721 | 27 | 13 | 56 |
Total number of gains: 109; Total number of losses: 55; Total Number of normals: 324.
Somatic mutations of PSME2:
Generating mutation plots.
Highly correlated genes for PSME2:
Showing top 20/190 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PSME2 | PSME1 | 0.795471 | 3 | 0 | 3 |
| PSME2 | DTX3L | 0.76961 | 3 | 0 | 3 |
| PSME2 | STAT1 | 0.768402 | 4 | 0 | 4 |
| PSME2 | DNMT1 | 0.757667 | 3 | 0 | 3 |
| PSME2 | UBE2C | 0.754135 | 3 | 0 | 3 |
| PSME2 | TOP2A | 0.748604 | 3 | 0 | 3 |
| PSME2 | KIF20A | 0.738203 | 3 | 0 | 3 |
| PSME2 | CCNB2 | 0.735714 | 3 | 0 | 3 |
| PSME2 | HPRT1 | 0.72857 | 3 | 0 | 3 |
| PSME2 | ATAD2 | 0.727796 | 3 | 0 | 3 |
| PSME2 | CENPE | 0.726452 | 3 | 0 | 3 |
| PSME2 | CEP55 | 0.724105 | 3 | 0 | 3 |
| PSME2 | BIRC5 | 0.723002 | 3 | 0 | 3 |
| PSME2 | HJURP | 0.720108 | 3 | 0 | 3 |
| PSME2 | PARP14 | 0.717632 | 4 | 0 | 4 |
| PSME2 | LIMK1 | 0.716084 | 3 | 0 | 3 |
| PSME2 | AURKA | 0.715619 | 3 | 0 | 3 |
| PSME2 | CKAP2 | 0.714066 | 3 | 0 | 3 |
| PSME2 | ASPM | 0.713941 | 3 | 0 | 3 |
| PSME2 | AGRN | 0.712283 | 3 | 0 | 3 |
For details and further investigation, click here