| Full name: signal-induced proliferation-associated 1 | Alias Symbol: SPA1 | ||
| Type: protein-coding gene | Cytoband: 11q13.1 | ||
| Entrez ID: 6494 | HGNC ID: HGNC:10885 | Ensembl Gene: ENSG00000213445 | OMIM ID: 602180 |
| Drug and gene relationship at DGIdb | |||
SIPA1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04015 | Rap1 signaling pathway | |
| hsa04670 | Leukocyte transendothelial migration |
Expression of SIPA1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SIPA1 | 6494 | 204164_at | 0.4655 | 0.0983 | |
| GSE20347 | SIPA1 | 6494 | 204164_at | 0.0289 | 0.8373 | |
| GSE23400 | SIPA1 | 6494 | 204164_at | -0.0740 | 0.1123 | |
| GSE26886 | SIPA1 | 6494 | 204164_at | 0.2554 | 0.0529 | |
| GSE29001 | SIPA1 | 6494 | 204164_at | -0.0325 | 0.8811 | |
| GSE38129 | SIPA1 | 6494 | 204164_at | -0.1117 | 0.3012 | |
| GSE45670 | SIPA1 | 6494 | 204164_at | 0.2077 | 0.0172 | |
| GSE53622 | SIPA1 | 6494 | 39327 | -0.0795 | 0.2483 | |
| GSE53624 | SIPA1 | 6494 | 39327 | -0.3059 | 0.0000 | |
| GSE63941 | SIPA1 | 6494 | 204164_at | 0.8149 | 0.0310 | |
| GSE77861 | SIPA1 | 6494 | 204164_at | 0.0751 | 0.6697 | |
| GSE97050 | SIPA1 | 6494 | A_23_P127460 | 0.1319 | 0.7604 | |
| SRP007169 | SIPA1 | 6494 | RNAseq | -1.0030 | 0.0108 | |
| SRP008496 | SIPA1 | 6494 | RNAseq | -1.0490 | 0.0001 | |
| SRP064894 | SIPA1 | 6494 | RNAseq | 0.5044 | 0.0707 | |
| SRP133303 | SIPA1 | 6494 | RNAseq | -0.0587 | 0.8033 | |
| SRP159526 | SIPA1 | 6494 | RNAseq | -0.3555 | 0.1299 | |
| SRP193095 | SIPA1 | 6494 | RNAseq | 0.1142 | 0.5058 | |
| SRP219564 | SIPA1 | 6494 | RNAseq | -0.0225 | 0.9466 | |
| TCGA | SIPA1 | 6494 | RNAseq | 0.1071 | 0.1164 |
Upregulated datasets: 0; Downregulated datasets: 2.
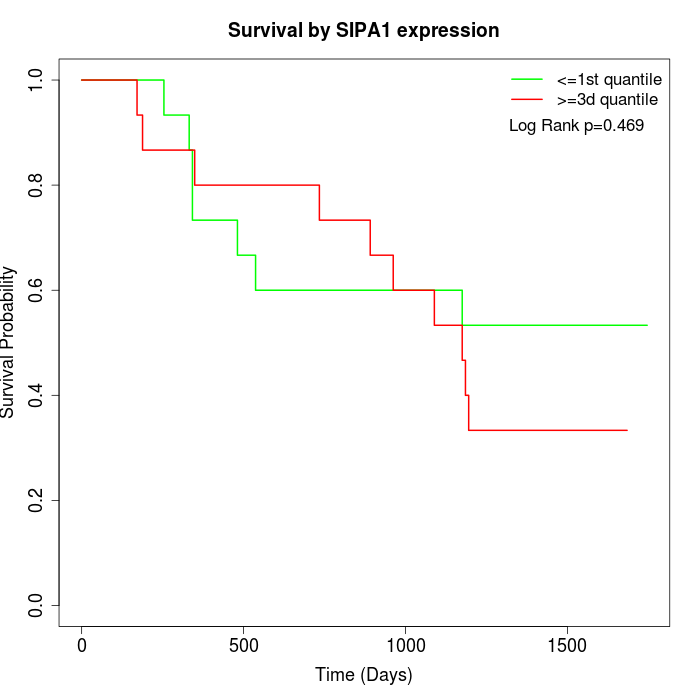
Survival by SIPA1 expression:
Note: Click image to view full size file.
Copy number change of SIPA1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SIPA1 | 6494 | 8 | 5 | 17 | |
| GSE20123 | SIPA1 | 6494 | 8 | 5 | 17 | |
| GSE43470 | SIPA1 | 6494 | 4 | 1 | 38 | |
| GSE46452 | SIPA1 | 6494 | 11 | 3 | 45 | |
| GSE47630 | SIPA1 | 6494 | 7 | 5 | 28 | |
| GSE54993 | SIPA1 | 6494 | 3 | 0 | 67 | |
| GSE54994 | SIPA1 | 6494 | 7 | 5 | 41 | |
| GSE60625 | SIPA1 | 6494 | 0 | 3 | 8 | |
| GSE74703 | SIPA1 | 6494 | 3 | 0 | 33 | |
| GSE74704 | SIPA1 | 6494 | 6 | 3 | 11 | |
| TCGA | SIPA1 | 6494 | 23 | 7 | 66 |
Total number of gains: 80; Total number of losses: 37; Total Number of normals: 371.
Somatic mutations of SIPA1:
Generating mutation plots.
Highly correlated genes for SIPA1:
Showing top 20/262 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SIPA1 | PPP1R9B | 0.738395 | 3 | 0 | 3 |
| SIPA1 | CD38 | 0.723711 | 3 | 0 | 3 |
| SIPA1 | KIF20A | 0.706096 | 3 | 0 | 3 |
| SIPA1 | SRC | 0.695687 | 4 | 0 | 4 |
| SIPA1 | TSPAN17 | 0.695029 | 3 | 0 | 3 |
| SIPA1 | CCDC86 | 0.690484 | 3 | 0 | 3 |
| SIPA1 | INO80B | 0.689272 | 4 | 0 | 4 |
| SIPA1 | ADAP2 | 0.671192 | 3 | 0 | 3 |
| SIPA1 | FAM111B | 0.666524 | 3 | 0 | 3 |
| SIPA1 | NCEH1 | 0.665766 | 3 | 0 | 3 |
| SIPA1 | LDHAL6B | 0.664586 | 3 | 0 | 3 |
| SIPA1 | HYOU1 | 0.656709 | 4 | 0 | 4 |
| SIPA1 | KIR2DL2 | 0.655737 | 3 | 0 | 3 |
| SIPA1 | TRIP13 | 0.651767 | 4 | 0 | 3 |
| SIPA1 | CPNE2 | 0.651664 | 5 | 0 | 5 |
| SIPA1 | SCAMP4 | 0.648284 | 4 | 0 | 3 |
| SIPA1 | ATXN7L3 | 0.643138 | 3 | 0 | 3 |
| SIPA1 | CTDNEP1 | 0.637219 | 5 | 0 | 4 |
| SIPA1 | INTS1 | 0.636671 | 4 | 0 | 4 |
| SIPA1 | MRPL36 | 0.633371 | 3 | 0 | 3 |
For details and further investigation, click here